Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1424-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1424-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 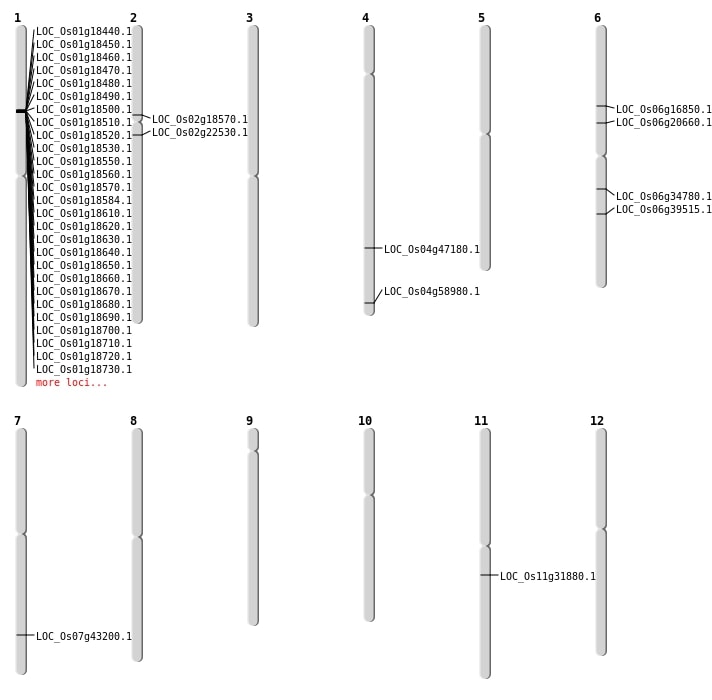
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17823737 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g32500 |
| SBS | Chr1 | 20080103 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 2382665 | A-G | HOMO | UTR_5_PRIME | |
| SBS | Chr1 | 25479624 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g44430 |
| SBS | Chr1 | 39038186 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12973451 | C-T | HET | INTRON | |
| SBS | Chr10 | 5304480 | C-T | HET | INTRON | |
| SBS | Chr10 | 6776389 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8668883 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 17902605 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18731003 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 23817490 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 24285165 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 27699841 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 10831972 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18570 |
| SBS | Chr2 | 11723886 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 13443025 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 28224967 | C-G | HET | UTR_5_PRIME | |
| SBS | Chr3 | 21858087 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 27780451 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 6493244 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 7798010 | T-C | HET | INTRON | |
| SBS | Chr4 | 15526245 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 17317021 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35088827 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g58980 |
| SBS | Chr5 | 24031616 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 23503381 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 3286456 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 3286457 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 11352522 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17885663 | A-T | HET | INTRON | |
| SBS | Chr7 | 25892024 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g43200 |
| SBS | Chr7 | 9835008 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 770796 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr8 | 9601478 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 16485323 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 9872912 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1037795 | 1037796 | 1 | |
| Deletion | Chr7 | 2198099 | 2198120 | 21 | |
| Deletion | Chr9 | 3491975 | 3491976 | 1 | |
| Deletion | Chr3 | 5363560 | 5363570 | 10 | |
| Deletion | Chr6 | 9759362 | 9759373 | 11 | LOC_Os06g16850 |
| Deletion | Chr1 | 10362395 | 11592486 | 1230091 | 175 |
| Deletion | Chr2 | 11311985 | 11311986 | 1 | |
| Deletion | Chr9 | 11757658 | 11757661 | 3 | |
| Deletion | Chr6 | 11905607 | 11905608 | 1 | LOC_Os06g20660 |
| Deletion | Chr2 | 11989684 | 11989693 | 9 | |
| Deletion | Chr11 | 12611093 | 12611094 | 1 | |
| Deletion | Chr1 | 12671405 | 12671663 | 258 | |
| Deletion | Chr2 | 13441569 | 13441571 | 2 | LOC_Os02g22530 |
| Deletion | Chr1 | 13998737 | 13998738 | 1 | |
| Deletion | Chr7 | 15343683 | 15343687 | 4 | |
| Deletion | Chr10 | 16382690 | 16382692 | 2 | |
| Deletion | Chr3 | 17957902 | 17957905 | 3 | |
| Deletion | Chr11 | 18732891 | 18732900 | 9 | LOC_Os11g31880 |
| Deletion | Chr12 | 24364974 | 24364975 | 1 | |
| Deletion | Chr11 | 25231409 | 25231426 | 17 | |
| Deletion | Chr4 | 28021576 | 28021580 | 4 | LOC_Os04g47180 |
| Deletion | Chr6 | 31078305 | 31078331 | 26 | |
| Deletion | Chr1 | 39829447 | 39830928 | 1481 | LOC_Os01g68570 |
| Deletion | Chr1 | 40603869 | 40603884 | 15 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 6636712 | 6636713 | 2 | |
| Insertion | Chr11 | 871553 | 871554 | 2 | |
| Insertion | Chr9 | 16220472 | 16220474 | 3 |
Inversions: 5
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 1959741 | Chr1 | 10109483 | LOC_Os02g04440 |
| Translocation | Chr2 | 1959752 | Chr1 | 10108411 | LOC_Os02g04440 |
| Translocation | Chr6 | 20213453 | Chr1 | 12671643 | 2 |
| Translocation | Chr6 | 20213454 | Chr1 | 12671434 | 2 |
| Translocation | Chr6 | 23463394 | Chr3 | 29226748 | LOC_Os06g39515 |
