Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1443-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1443-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 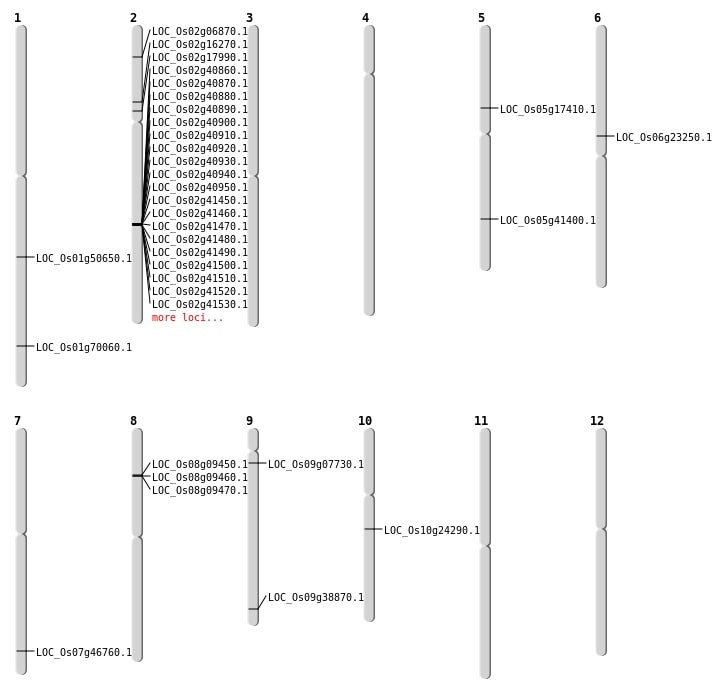
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22271234 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 25651825 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29094342 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g50650 |
| SBS | Chr10 | 12443024 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24290 |
| SBS | Chr10 | 17240761 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 18380439 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21013928 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 21013929 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 8388355 | T-A | HET | INTRON | |
| SBS | Chr11 | 14114199 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 28829005 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 7762528 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 10179181 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 13109040 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 24540561 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 23501150 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 26710475 | C-A | HET | INTRON | |
| SBS | Chr2 | 29612268 | T-A | HET | INTRON | |
| SBS | Chr2 | 33733056 | C-T | HET | STOP_GAINED | LOC_Os02g55070 |
| SBS | Chr2 | 9261449 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g16270 |
| SBS | Chr3 | 16746909 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 16746910 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 19003237 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 12000024 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18616703 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 27860890 | T-G | HET | INTRON | |
| SBS | Chr5 | 7679900 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13573246 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23250 |
| SBS | Chr6 | 15157477 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 15204492 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 15642525 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 16965851 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 23767627 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9367033 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 9445914 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 9445915 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 13669211 | C-T | HET | INTRON | |
| SBS | Chr7 | 16443538 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 26464165 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 8718991 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 22013514 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 26261737 | C-T | HET | INTRON | |
| SBS | Chr9 | 10313405 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1558067 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3899692 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07730 |
| SBS | Chr9 | 6735634 | G-C | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 3462559 | 3462563 | 4 | LOC_Os02g06870 |
| Deletion | Chr1 | 4368463 | 4368465 | 2 | |
| Deletion | Chr8 | 5477130 | 5489206 | 12076 | 3 |
| Deletion | Chr12 | 6950858 | 6950860 | 2 | |
| Deletion | Chr2 | 7047637 | 7047640 | 3 | |
| Deletion | Chr1 | 9472461 | 9472464 | 3 | |
| Deletion | Chr5 | 9995151 | 9995155 | 4 | LOC_Os05g17410 |
| Deletion | Chr2 | 10438804 | 10438811 | 7 | LOC_Os02g17990 |
| Deletion | Chr12 | 12739556 | 12739557 | 1 | |
| Deletion | Chr10 | 14550121 | 14550125 | 4 | |
| Deletion | Chr12 | 17274208 | 17274215 | 7 | |
| Deletion | Chr10 | 17894271 | 17894272 | 1 | |
| Deletion | Chr12 | 18067029 | 18067035 | 6 | |
| Deletion | Chr5 | 21761139 | 21761140 | 1 | |
| Deletion | Chr9 | 22327743 | 22327748 | 5 | LOC_Os09g38870 |
| Deletion | Chr6 | 23152789 | 23152792 | 3 | |
| Deletion | Chr5 | 24244843 | 24244846 | 3 | LOC_Os05g41400 |
| Deletion | Chr1 | 24313248 | 24313250 | 2 | |
| Deletion | Chr2 | 24767001 | 25188000 | 421000 | 55 |
| Deletion | Chr4 | 26745647 | 26745654 | 7 | |
| Deletion | Chr7 | 27947942 | 27947943 | 1 | LOC_Os07g46760 |
| Deletion | Chr5 | 28881468 | 28881470 | 2 | |
| Deletion | Chr2 | 32178360 | 32419952 | 241592 | 40 |
| Deletion | Chr1 | 40554645 | 40554646 | 1 | LOC_Os01g70060 |
| Deletion | Chr1 | 42135984 | 42135985 | 1 |
Insertions: 5
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3250052 | Chr3 | 24406169 |
