Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1463-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1463-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 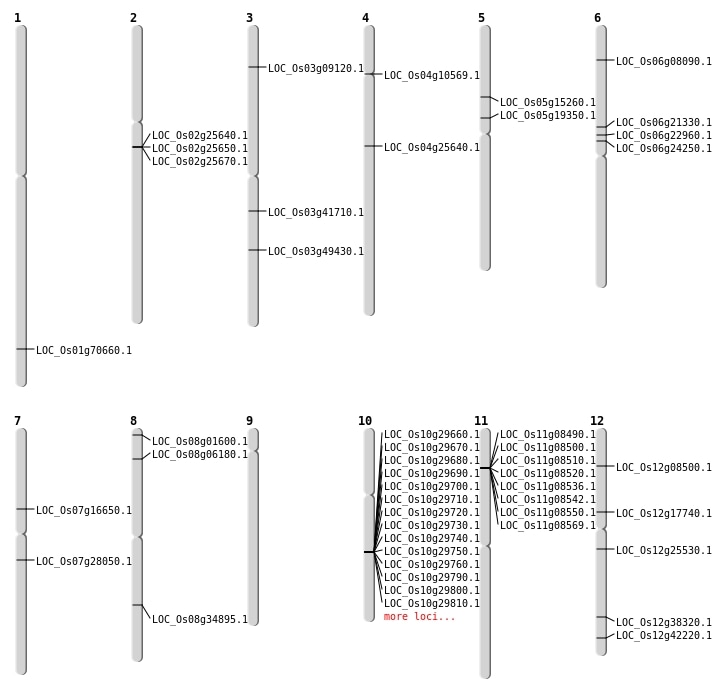
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1814856 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 2141408 | T-C | HOMO | INTRON | |
| SBS | Chr1 | 30877811 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 319576 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 39945586 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 10508642 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1823921 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1823922 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 10159735 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g17740 |
| SBS | Chr12 | 26106709 | A-G | HET | INTRON | |
| SBS | Chr12 | 4311891 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g08500 |
| SBS | Chr12 | 7933447 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9921801 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 10202659 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 33127137 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3978204 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14855947 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g25640 |
| SBS | Chr4 | 22724307 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 5739657 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g10569 |
| SBS | Chr4 | 7800916 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11237878 | C-T | HOMO | STOP_GAINED | LOC_Os05g19350 |
| SBS | Chr5 | 6519464 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 12318935 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g21330 |
| SBS | Chr7 | 18984209 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 6048921 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 7779782 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 9775462 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16650 |
| SBS | Chr8 | 12628816 | G-A | HET | INTRON | |
| SBS | Chr8 | 13034032 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 21974167 | G-A | HOMO | START_GAINED | LOC_Os08g34895 |
| SBS | Chr9 | 1069778 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 17474300 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9271519 | C-T | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1448857 | 1448860 | 3 | |
| Deletion | Chr8 | 3412758 | 3412771 | 13 | LOC_Os08g06180 |
| Deletion | Chr11 | 4483001 | 4525000 | 42000 | 8 |
| Deletion | Chr2 | 8368359 | 8368362 | 3 | |
| Deletion | Chr5 | 8655628 | 8655638 | 10 | LOC_Os05g15260 |
| Deletion | Chr9 | 9098577 | 9098580 | 3 | |
| Deletion | Chr11 | 9222409 | 9222437 | 28 | |
| Deletion | Chr11 | 10472843 | 10472845 | 2 | |
| Deletion | Chr4 | 13123690 | 13123691 | 1 | |
| Deletion | Chr12 | 14782058 | 14782062 | 4 | LOC_Os12g25530 |
| Deletion | Chr2 | 14986001 | 15027000 | 41000 | 3 |
| Deletion | Chr10 | 15427001 | 15925000 | 498000 | 84 |
| Deletion | Chr4 | 17198875 | 17198934 | 59 | |
| Deletion | Chr9 | 20256794 | 20256805 | 11 | |
| Deletion | Chr7 | 20384955 | 20384958 | 3 | |
| Deletion | Chr12 | 21146851 | 21146856 | 5 | |
| Deletion | Chr12 | 22531360 | 22531365 | 5 | LOC_Os12g36770 |
| Deletion | Chr3 | 23173222 | 23173231 | 9 | LOC_Os03g41710 |
| Deletion | Chr8 | 23176737 | 23176757 | 20 | |
| Deletion | Chr12 | 26194535 | 26194546 | 11 | LOC_Os12g42220 |
| Deletion | Chr3 | 28140939 | 28140943 | 4 | LOC_Os03g49430 |
| Deletion | Chr1 | 34576311 | 34576352 | 41 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 22223380 | 22223383 | 4 | |
| Insertion | Chr11 | 27699077 | 27699078 | 2 | |
| Insertion | Chr12 | 23521192 | 23521195 | 4 | LOC_Os12g38320 |
| Insertion | Chr2 | 23925763 | 23925763 | 1 | |
| Insertion | Chr2 | 27647359 | 27647360 | 2 | |
| Insertion | Chr3 | 21741766 | 21741767 | 2 | |
| Insertion | Chr4 | 16781947 | 16781948 | 2 | |
| Insertion | Chr7 | 8045906 | 8045909 | 4 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 3926660 | 3976479 | LOC_Os06g08090 |
| Inversion | Chr6 | 13176361 | 13408768 | LOC_Os06g22960 |
| Inversion | Chr6 | 13176381 | 13408784 | LOC_Os06g22960 |
| Inversion | Chr6 | 14198731 | 14454824 | LOC_Os06g24250 |
| Inversion | Chr10 | 15013277 | 15427224 | LOC_Os10g29660 |
| Inversion | Chr1 | 40907142 | 41174134 | LOC_Os01g70660 |
| Inversion | Chr1 | 40907164 | 41174159 | LOC_Os01g70660 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 360094 | Chr2 | 532399 | LOC_Os08g01600 |
| Translocation | Chr3 | 4749055 | Chr1 | 29064641 | LOC_Os03g09120 |
| Translocation | Chr8 | 18631943 | Chr7 | 16356425 | LOC_Os07g28050 |
