Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1466-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1466-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 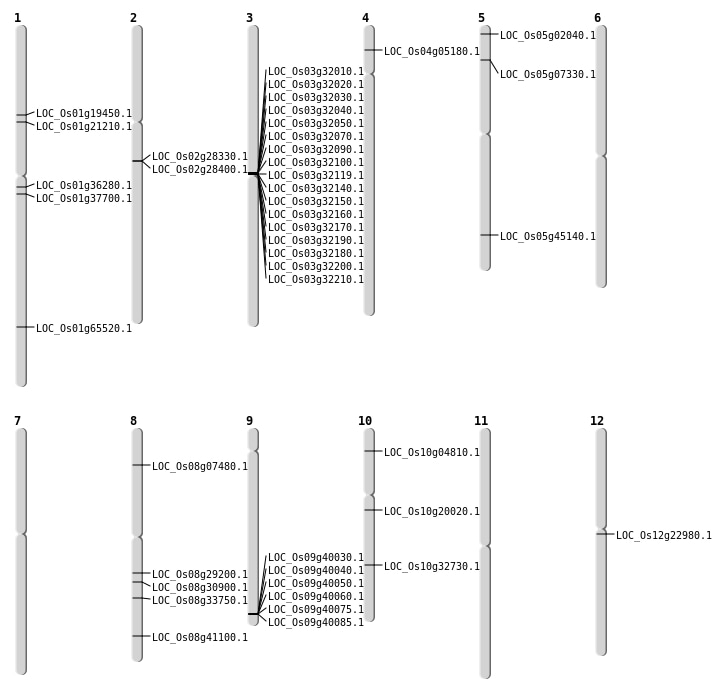
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11420025 | T-C | Homo | INTRON | |
| SBS | Chr1 | 15822758 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 2019320 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 28749998 | G-T | HET | INTRON | |
| SBS | Chr1 | 28750001 | A-G | HET | INTRON | |
| SBS | Chr10 | 10043080 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g20020 |
| SBS | Chr10 | 17118894 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 2484291 | G-A | Homo | INTRON | |
| SBS | Chr11 | 27698016 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 679034 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 13238514 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 23202868 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 32625278 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 9042493 | G-A | Homo | INTRON | |
| SBS | Chr3 | 25747445 | A-G | HET | INTRON | |
| SBS | Chr4 | 17184290 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 17576238 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 18614177 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2583469 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05180 |
| SBS | Chr4 | 27621532 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 3319164 | A-T | Homo | INTRON | |
| SBS | Chr5 | 3899709 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g07330 |
| SBS | Chr5 | 596373 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g02040 |
| SBS | Chr6 | 11942741 | G-A | HET | INTRON | |
| SBS | Chr7 | 28112975 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6785412 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 19072053 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30900 |
| SBS | Chr8 | 4197182 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g07480 |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2103035 | 2103037 | 2 | |
| Deletion | Chr10 | 2320214 | 2330155 | 9941 | LOC_Os10g04810 |
| Deletion | Chr9 | 2442093 | 2442094 | 1 | |
| Deletion | Chr5 | 5129820 | 5129821 | 1 | |
| Deletion | Chr7 | 5736665 | 5736671 | 6 | |
| Deletion | Chr10 | 6560824 | 6560825 | 1 | |
| Deletion | Chr1 | 11835729 | 11835755 | 26 | LOC_Os01g21210 |
| Deletion | Chr12 | 12991177 | 12991185 | 8 | LOC_Os12g22980 |
| Deletion | Chr2 | 13620939 | 13635289 | 14350 | |
| Deletion | Chr12 | 14113858 | 14113859 | 1 | |
| Deletion | Chr4 | 14128543 | 14128544 | 1 | |
| Deletion | Chr6 | 14925418 | 14925427 | 9 | |
| Deletion | Chr2 | 16743566 | 16744086 | 520 | LOC_Os02g28330 |
| Deletion | Chr2 | 16789973 | 16789982 | 9 | LOC_Os02g28400 |
| Deletion | Chr8 | 17905574 | 17905578 | 4 | LOC_Os08g29200 |
| Deletion | Chr3 | 18306001 | 18427000 | 121000 | 17 |
| Deletion | Chr3 | 20101450 | 20101451 | 1 | |
| Deletion | Chr3 | 20541202 | 20541203 | 1 | |
| Deletion | Chr10 | 22384013 | 22384025 | 12 | |
| Deletion | Chr10 | 22849413 | 22849414 | 1 | |
| Deletion | Chr9 | 22978001 | 23013000 | 35000 | 6 |
| Deletion | Chr8 | 23826109 | 23826390 | 281 | |
| Deletion | Chr8 | 23902321 | 23902327 | 6 | |
| Deletion | Chr7 | 24103192 | 24103198 | 6 | |
| Deletion | Chr11 | 24575358 | 24575482 | 124 | |
| Deletion | Chr3 | 24994608 | 24994610 | 2 | |
| Deletion | Chr3 | 25163323 | 25163328 | 5 | |
| Deletion | Chr8 | 25530398 | 25530400 | 2 | |
| Deletion | Chr11 | 25939426 | 25939445 | 19 | |
| Deletion | Chr8 | 25975756 | 25975783 | 27 | LOC_Os08g41100 |
| Deletion | Chr5 | 26212169 | 26212173 | 4 | LOC_Os05g45140 |
| Deletion | Chr7 | 26339271 | 26339341 | 70 | |
| Deletion | Chr2 | 28136309 | 28136328 | 19 | |
| Deletion | Chr6 | 30923107 | 30923108 | 1 | |
| Deletion | Chr1 | 41143680 | 41143683 | 3 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11005990 | 11005990 | 1 | LOC_Os01g19450 |
| Insertion | Chr1 | 28208037 | 28208037 | 1 | |
| Insertion | Chr1 | 37785629 | 37785629 | 1 | |
| Insertion | Chr10 | 12598029 | 12598089 | 61 | |
| Insertion | Chr11 | 11993691 | 11993691 | 1 | |
| Insertion | Chr12 | 4131798 | 4131813 | 16 | |
| Insertion | Chr4 | 25013786 | 25013817 | 32 | |
| Insertion | Chr5 | 12495322 | 12495322 | 1 | |
| Insertion | Chr6 | 5883097 | 5883097 | 1 | |
| Insertion | Chr7 | 4570092 | 4570093 | 2 | |
| Insertion | Chr8 | 21090259 | 21090262 | 4 | LOC_Os08g33750 |
| Insertion | Chr9 | 12637135 | 12637189 | 55 | |
| Insertion | Chr9 | 22367053 | 22367053 | 1 | |
| Insertion | Chr9 | 22463086 | 22463091 | 6 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 16993116 | 17133254 | LOC_Os10g32730 |
| Inversion | Chr1 | 20114074 | 21102014 | LOC_Os01g36280 |
| Inversion | Chr1 | 20114097 | 21078679 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 14855608 | Chr1 | 38029221 | LOC_Os01g65520 |
| Translocation | Chr9 | 14855649 | Chr1 | 38028443 | LOC_Os01g65520 |
