Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1477-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1477-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 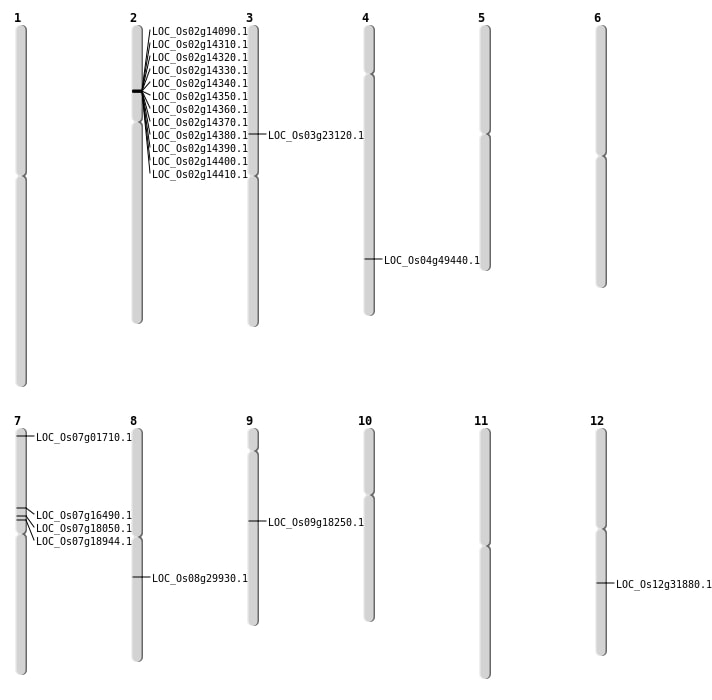
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22124108 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 24507212 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 26571718 | G-C | HET | INTRON | |
| SBS | Chr1 | 334586 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 42052750 | T-G | Homo | INTERGENIC | |
| SBS | Chr10 | 17579370 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3023243 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 5500939 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 24681104 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 11863417 | A-T | HET | INTRON | |
| SBS | Chr12 | 19201403 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g31880 |
| SBS | Chr12 | 7205890 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 12497959 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 2044212 | C-A | Homo | INTERGENIC | |
| SBS | Chr3 | 1131803 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 13370559 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23120 |
| SBS | Chr3 | 13809827 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 31863571 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 3696708 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr4 | 11117085 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 14622081 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 26187380 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 3906719 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 10285990 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 12524160 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 19695267 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 28349841 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9254972 | A-G | HET | INTRON | |
| SBS | Chr7 | 9663993 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16490 |
| SBS | Chr8 | 13046817 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17414417 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 2656931 | T-G | Homo | INTERGENIC | |
| SBS | Chr8 | 8727974 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8212198 | A-G | HET | INTERGENIC |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 47109 | 47110 | 1 | |
| Deletion | Chr7 | 427694 | 427695 | 1 | LOC_Os07g01710 |
| Deletion | Chr7 | 1431570 | 1431572 | 2 | |
| Deletion | Chr10 | 1519144 | 1519454 | 310 | |
| Deletion | Chr10 | 2402062 | 2402065 | 3 | |
| Deletion | Chr10 | 4069573 | 4069579 | 6 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr12 | 5668416 | 5668418 | 2 | |
| Deletion | Chr2 | 6352472 | 6352474 | 2 | |
| Deletion | Chr1 | 6400198 | 6400206 | 8 | |
| Deletion | Chr11 | 7286199 | 7286200 | 1 | |
| Deletion | Chr10 | 7448387 | 7448405 | 18 | |
| Deletion | Chr9 | 7691761 | 7691795 | 34 | |
| Deletion | Chr2 | 7855001 | 7916000 | 61000 | 11 |
| Deletion | Chr3 | 7943470 | 7943472 | 2 | |
| Deletion | Chr7 | 10101396 | 10101397 | 1 | |
| Deletion | Chr1 | 10252588 | 10252830 | 242 | |
| Deletion | Chr7 | 10683086 | 10683087 | 1 | LOC_Os07g18050 |
| Deletion | Chr9 | 11201987 | 11201993 | 6 | LOC_Os09g18250 |
| Deletion | Chr7 | 11222425 | 11222426 | 1 | LOC_Os07g18944 |
| Deletion | Chr12 | 11499010 | 11499031 | 21 | |
| Deletion | Chr10 | 11565194 | 11565199 | 5 | |
| Deletion | Chr5 | 11995520 | 11995530 | 10 | |
| Deletion | Chr11 | 12058162 | 12058196 | 34 | |
| Deletion | Chr4 | 13854628 | 13854649 | 21 | |
| Deletion | Chr3 | 14665187 | 14665188 | 1 | |
| Deletion | Chr9 | 16323601 | 16323608 | 7 | |
| Deletion | Chr2 | 16463751 | 16463803 | 52 | |
| Deletion | Chr5 | 17465211 | 17465213 | 2 | |
| Deletion | Chr8 | 18386255 | 18386256 | 1 | LOC_Os08g29930 |
| Deletion | Chr6 | 22262829 | 22262830 | 1 | |
| Deletion | Chr3 | 22468508 | 22468573 | 65 | |
| Deletion | Chr5 | 24448166 | 24448167 | 1 | |
| Deletion | Chr12 | 24525783 | 24525805 | 22 | |
| Deletion | Chr2 | 25204499 | 25204500 | 1 | |
| Deletion | Chr8 | 25436797 | 25436807 | 10 | |
| Deletion | Chr2 | 25567677 | 25567678 | 1 | |
| Deletion | Chr6 | 25662514 | 25662515 | 1 | |
| Deletion | Chr4 | 26648657 | 26648700 | 43 | |
| Deletion | Chr5 | 26970125 | 26970130 | 5 | |
| Deletion | Chr4 | 29500341 | 29500359 | 18 | LOC_Os04g49440 |
Insertions: 13
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6641078 | 6655885 | |
| Inversion | Chr2 | 7690952 | 8540748 | LOC_Os02g14090 |
| Inversion | Chr2 | 7854849 | 8540747 | |
| Inversion | Chr2 | 7915543 | 7926291 | LOC_Os02g14410 |
| Inversion | Chr1 | 20459422 | 22117876 | |
| Inversion | Chr5 | 24965409 | 25059780 |
No Translocation
