Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1488-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1488-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 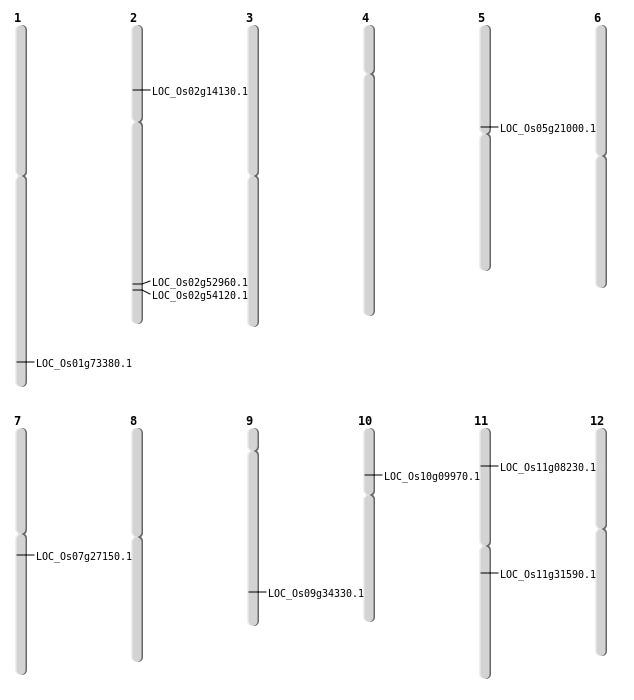
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21742558 | C-T | HET | INTRON | |
| SBS | Chr1 | 32952566 | T-A | Homo | INTERGENIC | |
| SBS | Chr10 | 11351597 | G-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 2008946 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 5407042 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g09970 |
| SBS | Chr11 | 10845900 | C-T | Homo | INTRON | |
| SBS | Chr11 | 4321471 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08230 |
| SBS | Chr12 | 21300383 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 6566532 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 12263984 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 24018353 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 25130036 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 25130038 | T-A | Homo | INTERGENIC | |
| SBS | Chr2 | 26939545 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 27609006 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 17866126 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 20053998 | C-T | Homo | INTRON | |
| SBS | Chr3 | 2100867 | T-A | Homo | SPLICE_SITE_REGION | |
| SBS | Chr4 | 1312023 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 27866918 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 12270422 | C-A | Homo | INTERGENIC | |
| SBS | Chr5 | 17176745 | C-A | Homo | UTR_3_PRIME | |
| SBS | Chr6 | 13420714 | C-T | HET | INTRON | |
| SBS | Chr7 | 13285150 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13687595 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 19384498 | C-T | Homo | INTRON | |
| SBS | Chr7 | 2058107 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 13742031 | G-T | Homo | INTRON | |
| SBS | Chr8 | 13742032 | A-T | Homo | INTRON | |
| SBS | Chr8 | 14146283 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 20269090 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g34330 |
| SBS | Chr9 | 3643453 | A-G | HET | INTERGENIC |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 628755 | 628790 | 35 | |
| Deletion | Chr11 | 1483556 | 1483557 | 1 | |
| Deletion | Chr11 | 1498369 | 1498370 | 1 | |
| Deletion | Chr10 | 2285838 | 2287809 | 1971 | |
| Deletion | Chr11 | 3793275 | 3793299 | 24 | |
| Deletion | Chr4 | 4281657 | 4281665 | 8 | |
| Deletion | Chr11 | 5476122 | 5476146 | 24 | |
| Deletion | Chr7 | 6669607 | 6669608 | 1 | |
| Deletion | Chr12 | 8238326 | 8238327 | 1 | |
| Deletion | Chr6 | 8349815 | 8349829 | 14 | |
| Deletion | Chr12 | 8602897 | 8602898 | 1 | |
| Deletion | Chr9 | 9058670 | 9058809 | 139 | |
| Deletion | Chr9 | 10582264 | 10582269 | 5 | |
| Deletion | Chr9 | 10879365 | 10879402 | 37 | |
| Deletion | Chr5 | 12356799 | 12356800 | 1 | LOC_Os05g21000 |
| Deletion | Chr11 | 14054829 | 14054830 | 1 | |
| Deletion | Chr7 | 14673526 | 14673528 | 2 | |
| Deletion | Chr7 | 15755722 | 15755724 | 2 | LOC_Os07g27150 |
| Deletion | Chr7 | 16353850 | 16353852 | 2 | |
| Deletion | Chr11 | 16402986 | 16402988 | 2 | |
| Deletion | Chr3 | 17866283 | 17869259 | 2976 | |
| Deletion | Chr3 | 18392873 | 18392874 | 1 | |
| Deletion | Chr2 | 18561505 | 18561518 | 13 | |
| Deletion | Chr3 | 19961395 | 19961399 | 4 | |
| Deletion | Chr12 | 20268940 | 20268942 | 2 | |
| Deletion | Chr12 | 20353570 | 20353583 | 13 | |
| Deletion | Chr5 | 21073837 | 21073838 | 1 | |
| Deletion | Chr7 | 21881014 | 21881015 | 1 | |
| Deletion | Chr6 | 23217509 | 23217510 | 1 | |
| Deletion | Chr12 | 23735696 | 23735729 | 33 | |
| Deletion | Chr12 | 24215377 | 24215379 | 2 | |
| Deletion | Chr6 | 26102884 | 26102887 | 3 | |
| Deletion | Chr11 | 26394514 | 26394515 | 1 | |
| Deletion | Chr11 | 26590995 | 26590996 | 1 | |
| Deletion | Chr2 | 27055578 | 27055580 | 2 | |
| Deletion | Chr5 | 27160672 | 27160694 | 22 | |
| Deletion | Chr2 | 27516494 | 27516496 | 2 | |
| Deletion | Chr3 | 28075037 | 28075039 | 2 | |
| Deletion | Chr2 | 28400932 | 28400934 | 2 | |
| Deletion | Chr5 | 29090880 | 29090891 | 11 | |
| Deletion | Chr1 | 32220399 | 32220401 | 2 | |
| Deletion | Chr2 | 32391894 | 32391895 | 1 | LOC_Os02g52960 |
| Deletion | Chr2 | 33160894 | 33160914 | 20 | LOC_Os02g54120 |
| Deletion | Chr1 | 39497259 | 39497262 | 3 | |
| Deletion | Chr1 | 42527690 | 42532115 | 4425 | LOC_Os01g73380 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4240252 | 4240255 | 4 | |
| Insertion | Chr1 | 4810002 | 4810002 | 1 | |
| Insertion | Chr11 | 13608666 | 13608666 | 1 | |
| Insertion | Chr11 | 18465026 | 18465068 | 43 | LOC_Os11g31590 |
| Insertion | Chr2 | 6051807 | 6051807 | 1 | |
| Insertion | Chr2 | 7719174 | 7719174 | 1 | LOC_Os02g14130 |
| Insertion | Chr3 | 15061050 | 15061050 | 1 | |
| Insertion | Chr3 | 22910304 | 22910309 | 6 | |
| Insertion | Chr4 | 18330057 | 18330068 | 12 | |
| Insertion | Chr4 | 3988999 | 3988999 | 1 | |
| Insertion | Chr5 | 194174 | 194177 | 4 | |
| Insertion | Chr5 | 29411337 | 29411378 | 42 |
No Inversion
No Translocation
