Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1512-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1512-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 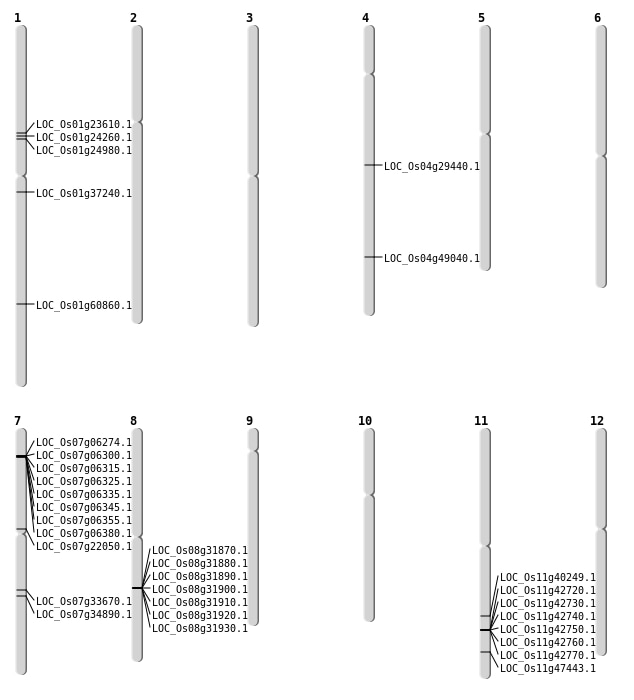
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10148008 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 20847287 | C-T | HET | INTRON | |
| SBS | Chr1 | 27416089 | C-G | Homo | INTERGENIC | |
| SBS | Chr1 | 32570228 | A-G | Homo | INTERGENIC | |
| SBS | Chr1 | 3960712 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 3967795 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10338458 | T-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr11 | 10339786 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 18622476 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 24007329 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g40249 |
| SBS | Chr11 | 28995722 | T-C | HET | INTRON | |
| SBS | Chr12 | 12677782 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 11770611 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 11770634 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 23761073 | A-T | Homo | INTERGENIC | |
| SBS | Chr2 | 27115408 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 34049971 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 1038935 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 346461 | C-T | Homo | INTRON | |
| SBS | Chr4 | 12407648 | A-G | HET | INTRON | |
| SBS | Chr4 | 17503578 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g29440 |
| SBS | Chr4 | 26806501 | C-G | HET | UTR_5_PRIME | |
| SBS | Chr4 | 29231856 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g49040 |
| SBS | Chr4 | 29231874 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g49040 |
| SBS | Chr5 | 20022563 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 20352628 | C-G | Homo | INTERGENIC | |
| SBS | Chr5 | 5767505 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 16595957 | T-A | Homo | INTRON | |
| SBS | Chr6 | 20565593 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 27824822 | A-G | HET | INTRON | |
| SBS | Chr6 | 30380626 | T-A | HET | INTRON | |
| SBS | Chr6 | 31199284 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 12326589 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g22050 |
| SBS | Chr7 | 13176159 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21347112 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 21365721 | G-A | HET | INTRON | |
| SBS | Chr7 | 23382986 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 13612653 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 17647013 | T-C | Homo | INTERGENIC | |
| SBS | Chr8 | 27417099 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 16211592 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5495925 | A-T | HET | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 863078 | 863084 | 6 | |
| Deletion | Chr9 | 1682579 | 1682580 | 1 | |
| Deletion | Chr2 | 1894936 | 1894939 | 3 | |
| Deletion | Chr7 | 3055001 | 3105000 | 50000 | 8 |
| Deletion | Chr12 | 4406968 | 4406973 | 5 | |
| Deletion | Chr10 | 5320508 | 5320509 | 1 | |
| Deletion | Chr1 | 8029594 | 8029615 | 21 | |
| Deletion | Chr12 | 9213418 | 9213420 | 2 | |
| Deletion | Chr9 | 10399185 | 10399186 | 1 | |
| Deletion | Chr11 | 11149105 | 11149120 | 15 | |
| Deletion | Chr11 | 11769443 | 11769444 | 1 | |
| Deletion | Chr6 | 14916251 | 14916253 | 2 | |
| Deletion | Chr11 | 16970635 | 16970641 | 6 | |
| Deletion | Chr9 | 17644037 | 17644046 | 9 | |
| Deletion | Chr3 | 18733812 | 18733814 | 2 | |
| Deletion | Chr10 | 19465898 | 19465903 | 5 | |
| Deletion | Chr2 | 19568211 | 19568216 | 5 | |
| Deletion | Chr8 | 19749001 | 19798000 | 49000 | 7 |
| Deletion | Chr7 | 19847195 | 19847196 | 1 | |
| Deletion | Chr6 | 19914425 | 19914426 | 1 | |
| Deletion | Chr8 | 20325970 | 20325971 | 1 | |
| Deletion | Chr1 | 20799529 | 20799539 | 10 | LOC_Os01g37240 |
| Deletion | Chr3 | 21149145 | 21149150 | 5 | |
| Deletion | Chr9 | 21756106 | 21756112 | 6 | |
| Deletion | Chr11 | 25735787 | 25761999 | 26212 | 6 |
| Deletion | Chr4 | 26408317 | 26408330 | 13 | |
| Deletion | Chr4 | 26627926 | 26627929 | 3 | |
| Deletion | Chr5 | 26892035 | 26892053 | 18 | |
| Deletion | Chr11 | 28621734 | 28621735 | 1 | LOC_Os11g47443 |
| Deletion | Chr11 | 28884331 | 28884334 | 3 | |
| Deletion | Chr2 | 33059367 | 33059389 | 22 | |
| Deletion | Chr1 | 35186043 | 35186044 | 1 | LOC_Os01g60860 |
| Deletion | Chr1 | 40330953 | 40330954 | 1 |
Insertions: 14
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 1797843 | 1860065 | |
| Inversion | Chr1 | 13100576 | 14544224 | |
| Inversion | Chr1 | 13281792 | 14080273 | 2 |
| Inversion | Chr1 | 13667268 | 14544215 | LOC_Os01g24260 |
| Inversion | Chr7 | 20116368 | 20912768 | 2 |
| Inversion | Chr3 | 26394529 | 27056321 |
No Translocation
