Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1514-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1514-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 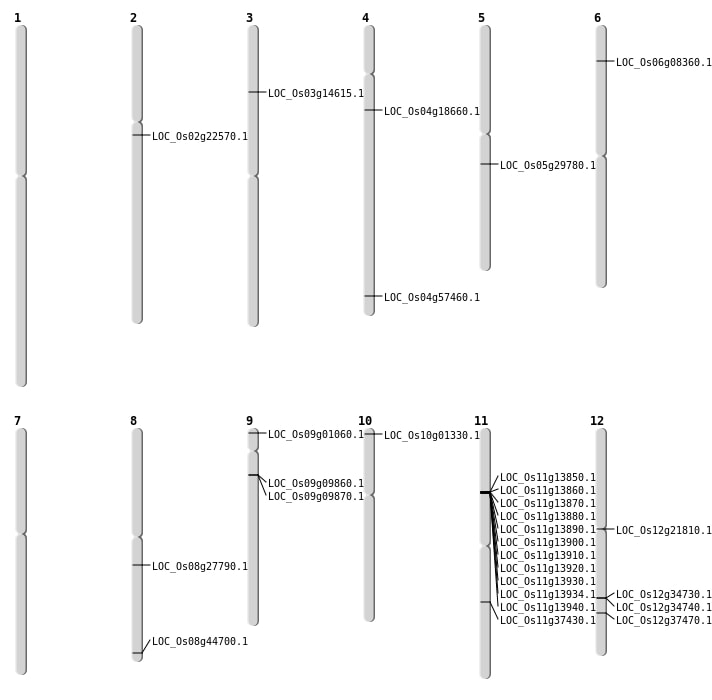
|
| Variant Information |
|---|
Single base substitutions: 54
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 8090802 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 16607758 | C-A | Homo | INTERGENIC | |
| SBS | Chr10 | 201534 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g01330 |
| SBS | Chr11 | 15990743 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 16140202 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 3274761 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 22997301 | C-T | HET | STOP_GAINED | LOC_Os12g37470 |
| SBS | Chr12 | 25546543 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26527945 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26846070 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 27201087 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 3881073 | G-A | Homo | INTRON | |
| SBS | Chr12 | 5752453 | T-C | HET | INTRON | |
| SBS | Chr12 | 9469429 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 14922047 | A-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr2 | 18112816 | A-G | Homo | INTRON | |
| SBS | Chr2 | 23553266 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 29823842 | T-C | HET | INTRON | |
| SBS | Chr2 | 35475567 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 35773606 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14961494 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 17907389 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19117932 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 2813158 | G-A | HET | INTRON | |
| SBS | Chr3 | 30997574 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 5048986 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 5048988 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr3 | 5056695 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 14317018 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 31587898 | C-T | HET | INTRON | |
| SBS | Chr4 | 33457528 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34183835 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g57460 |
| SBS | Chr4 | 4747211 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 970506 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 12672722 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 1698863 | A-C | Homo | INTERGENIC | |
| SBS | Chr6 | 17652576 | T-A | Homo | INTERGENIC | |
| SBS | Chr6 | 3361757 | C-T | HET | INTRON | |
| SBS | Chr7 | 11926411 | C-T | HET | INTRON | |
| SBS | Chr7 | 15956030 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 1885820 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 8437178 | G-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 28288483 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 6205969 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr9 | 13805119 | C-G | HET | INTRON | |
| SBS | Chr9 | 1385011 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 18604632 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 1922593 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 20875543 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 5356654 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09860 |
| SBS | Chr9 | 5356657 | C-T | HET | STOP_GAINED | LOC_Os09g09860 |
| SBS | Chr9 | 6341862 | T-G | HET | INTRON | |
| SBS | Chr9 | 7847293 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 88258 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01060 |
Deletions: 49
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 873113 | 873117 | 4 | |
| Deletion | Chr4 | 1191208 | 1191222 | 14 | |
| Deletion | Chr6 | 4063511 | 4063548 | 37 | LOC_Os06g08360 |
| Deletion | Chr3 | 5018492 | 5018497 | 5 | |
| Deletion | Chr4 | 5021409 | 5021411 | 2 | |
| Deletion | Chr9 | 5364586 | 5364605 | 19 | LOC_Os09g09870 |
| Deletion | Chr3 | 6623928 | 6623930 | 2 | |
| Deletion | Chr11 | 7643001 | 7720000 | 77000 | 10 |
| Deletion | Chr5 | 9181533 | 9181534 | 1 | |
| Deletion | Chr7 | 9457039 | 9457040 | 1 | |
| Deletion | Chr9 | 9964634 | 9964637 | 3 | |
| Deletion | Chr11 | 10630214 | 10630219 | 5 | |
| Deletion | Chr6 | 11088113 | 11088114 | 1 | |
| Deletion | Chr12 | 11379066 | 11379067 | 1 | |
| Deletion | Chr12 | 11622133 | 11622138 | 5 | |
| Deletion | Chr12 | 11637665 | 11637666 | 1 | |
| Deletion | Chr12 | 12276598 | 12276617 | 19 | LOC_Os12g21810 |
| Deletion | Chr11 | 12315140 | 12315295 | 155 | |
| Deletion | Chr1 | 12945458 | 12945460 | 2 | |
| Deletion | Chr3 | 13585056 | 13585057 | 1 | |
| Deletion | Chr8 | 13758559 | 13758560 | 1 | |
| Deletion | Chr1 | 13837859 | 13837861 | 2 | |
| Deletion | Chr3 | 14274349 | 14274350 | 1 | |
| Deletion | Chr8 | 14433029 | 14433063 | 34 | |
| Deletion | Chr3 | 14614914 | 14614920 | 6 | |
| Deletion | Chr4 | 14926925 | 14926939 | 14 | |
| Deletion | Chr2 | 15982298 | 15984167 | 1869 | |
| Deletion | Chr12 | 16027208 | 16027209 | 1 | |
| Deletion | Chr9 | 16562308 | 16562309 | 1 | |
| Deletion | Chr11 | 16684297 | 16684331 | 34 | |
| Deletion | Chr12 | 16895019 | 16895029 | 10 | |
| Deletion | Chr8 | 16925123 | 16928391 | 3268 | LOC_Os08g27790 |
| Deletion | Chr10 | 17119661 | 17119685 | 24 | |
| Deletion | Chr5 | 17228704 | 17234161 | 5457 | LOC_Os05g29780 |
| Deletion | Chr12 | 17461941 | 17461942 | 1 | |
| Deletion | Chr8 | 17964425 | 17964451 | 26 | |
| Deletion | Chr6 | 19659024 | 19659025 | 1 | |
| Deletion | Chr2 | 20696562 | 20696570 | 8 | |
| Deletion | Chr10 | 21348831 | 21348849 | 18 | |
| Deletion | Chr4 | 21582900 | 21582916 | 16 | |
| Deletion | Chr7 | 21698685 | 21698693 | 8 | |
| Deletion | Chr5 | 22701209 | 22701213 | 4 | |
| Deletion | Chr7 | 23847848 | 23847855 | 7 | |
| Deletion | Chr5 | 25428218 | 25428219 | 1 | |
| Deletion | Chr12 | 25521561 | 25521562 | 1 | |
| Deletion | Chr4 | 27050849 | 27050852 | 3 | |
| Deletion | Chr8 | 28094716 | 28094728 | 12 | LOC_Os08g44700 |
| Deletion | Chr5 | 28297593 | 28297594 | 1 | |
| Deletion | Chr2 | 29803973 | 29803984 | 11 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 30684929 | 30684930 | 2 | |
| Insertion | Chr1 | 34456218 | 34456218 | 1 | |
| Insertion | Chr10 | 16226519 | 16226520 | 2 | |
| Insertion | Chr10 | 20546495 | 20546495 | 1 | |
| Insertion | Chr11 | 22107950 | 22107955 | 6 | LOC_Os11g37430 |
| Insertion | Chr2 | 17832814 | 17832828 | 15 | |
| Insertion | Chr3 | 1263308 | 1263308 | 1 | |
| Insertion | Chr3 | 19575362 | 19575439 | 78 | |
| Insertion | Chr3 | 6553764 | 6553773 | 10 | |
| Insertion | Chr3 | 7939365 | 7939373 | 9 | LOC_Os03g14615 |
| Insertion | Chr4 | 5472791 | 5472791 | 1 | |
| Insertion | Chr6 | 22182993 | 22183001 | 9 | |
| Insertion | Chr7 | 12925214 | 12925235 | 22 | |
| Insertion | Chr7 | 18898804 | 18898804 | 1 | |
| Insertion | Chr8 | 24481453 | 24481482 | 30 | |
| Insertion | Chr8 | 2884665 | 2884665 | 1 | |
| Insertion | Chr9 | 15853656 | 15853703 | 48 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 7213520 | 7720228 | |
| Inversion | Chr2 | 13416055 | 13456470 | LOC_Os02g22570 |
| Inversion | Chr12 | 17346668 | 17346926 | |
| Inversion | Chr5 | 25529331 | 25535980 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 7213509 | Chr2 | 13371802 | |
| Translocation | Chr11 | 7427059 | Chr2 | 15984201 | |
| Translocation | Chr11 | 7642485 | Chr2 | 13371806 | LOC_Os11g13850 |
| Translocation | Chr12 | 21090321 | Chr4 | 10335314 | 2 |
| Translocation | Chr12 | 21094577 | Chr4 | 10335311 | 2 |
| Translocation | Chr12 | 21094717 | Chr7 | 24651099 | LOC_Os12g34740 |
| Translocation | Chr12 | 21094718 | Chr7 | 24651100 | LOC_Os12g34740 |
