Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1538-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1538-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 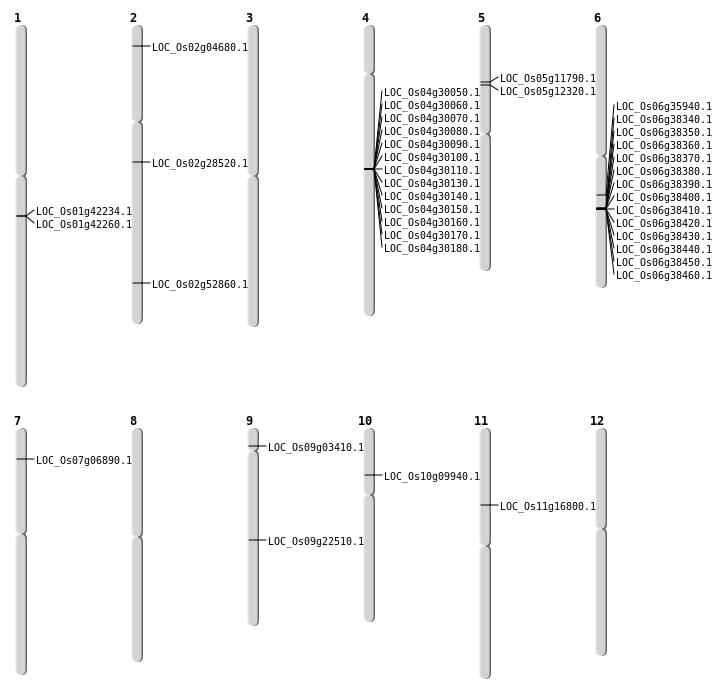
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10637884 | C-A | Homo | INTERGENIC | |
| SBS | Chr1 | 23977260 | T-C | HET | INTRON | |
| SBS | Chr1 | 26479235 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 15068091 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 4878777 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 5898301 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 15816209 | G-A | Homo | INTRON | |
| SBS | Chr11 | 24871543 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr11 | 9453563 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 24251601 | G-C | Homo | INTRON | |
| SBS | Chr12 | 9864108 | C-A | Homo | INTERGENIC | |
| SBS | Chr2 | 23478053 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 29134244 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 8152735 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19081260 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 25577455 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34371142 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 35013132 | A-G | HET | INTRON | |
| SBS | Chr3 | 4881196 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 10477968 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 5878648 | C-A | Homo | INTERGENIC | |
| SBS | Chr6 | 20471084 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 22681695 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5405356 | T-C | Homo | INTRON | |
| SBS | Chr6 | 5407426 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 3369605 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g06890 |
| SBS | Chr8 | 16743012 | T-A | Homo | INTERGENIC | |
| SBS | Chr8 | 19854187 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 7381911 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1684862 | A-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1684863 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g03410 |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 642590 | 642598 | 8 | |
| Deletion | Chr8 | 987974 | 987976 | 2 | |
| Deletion | Chr10 | 2199069 | 2199077 | 8 | |
| Deletion | Chr6 | 2755843 | 2755845 | 2 | |
| Deletion | Chr4 | 3404302 | 3404307 | 5 | |
| Deletion | Chr10 | 3846194 | 3846196 | 2 | |
| Deletion | Chr2 | 4272364 | 4272365 | 1 | |
| Deletion | Chr10 | 5386645 | 5386651 | 6 | LOC_Os10g09940 |
| Deletion | Chr10 | 5719152 | 5719153 | 1 | |
| Deletion | Chr2 | 7718685 | 7718690 | 5 | |
| Deletion | Chr8 | 8652050 | 8652130 | 80 | |
| Deletion | Chr11 | 9328462 | 9328465 | 3 | LOC_Os11g16800 |
| Deletion | Chr3 | 11036709 | 11036712 | 3 | |
| Deletion | Chr1 | 12097019 | 12097025 | 6 | |
| Deletion | Chr9 | 12576803 | 12576816 | 13 | |
| Deletion | Chr5 | 12698620 | 12698625 | 5 | |
| Deletion | Chr8 | 12707210 | 12707211 | 1 | |
| Deletion | Chr9 | 13581219 | 13581221 | 2 | LOC_Os09g22510 |
| Deletion | Chr4 | 15495314 | 15495327 | 13 | |
| Deletion | Chr2 | 16873953 | 16879577 | 5624 | LOC_Os02g28520 |
| Deletion | Chr9 | 17849630 | 17849647 | 17 | |
| Deletion | Chr4 | 17931001 | 18014000 | 83000 | 14 |
| Deletion | Chr9 | 18205026 | 18205036 | 10 | |
| Deletion | Chr4 | 18352594 | 18352596 | 2 | |
| Deletion | Chr9 | 20618435 | 20618444 | 9 | |
| Deletion | Chr6 | 20971837 | 20971861 | 24 | LOC_Os06g35940 |
| Deletion | Chr12 | 21510886 | 21510887 | 1 | |
| Deletion | Chr4 | 21637706 | 21637717 | 11 | |
| Deletion | Chr8 | 21900832 | 21900890 | 58 | |
| Deletion | Chr10 | 22588135 | 22588137 | 2 | |
| Deletion | Chr6 | 22711001 | 22794000 | 83000 | 13 |
| Deletion | Chr7 | 22869154 | 22869155 | 1 | |
| Deletion | Chr1 | 23932390 | 23941815 | 9425 | 2 |
| Deletion | Chr11 | 25198361 | 25198362 | 1 | |
| Deletion | Chr3 | 26887050 | 26887051 | 1 | |
| Deletion | Chr8 | 27263915 | 27263916 | 1 | |
| Deletion | Chr1 | 27829075 | 27829079 | 4 | |
| Deletion | Chr5 | 28248265 | 28248268 | 3 | |
| Deletion | Chr4 | 30875124 | 30875125 | 1 | |
| Deletion | Chr1 | 32205841 | 32205842 | 1 | |
| Deletion | Chr2 | 32323204 | 32323208 | 4 | LOC_Os02g52860 |
| Deletion | Chr4 | 33808242 | 33808244 | 2 | |
| Deletion | Chr3 | 34885572 | 34885582 | 10 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20121555 | 20121555 | 1 | |
| Insertion | Chr10 | 2919680 | 2919680 | 1 | |
| Insertion | Chr11 | 26799973 | 26799974 | 2 | |
| Insertion | Chr11 | 6666254 | 6666254 | 1 | |
| Insertion | Chr12 | 23039621 | 23039665 | 45 | |
| Insertion | Chr12 | 6752095 | 6752111 | 17 | |
| Insertion | Chr2 | 2106504 | 2106505 | 2 | LOC_Os02g04680 |
| Insertion | Chr2 | 33006906 | 33006907 | 2 | |
| Insertion | Chr2 | 35175745 | 35175746 | 2 | |
| Insertion | Chr3 | 18673295 | 18673295 | 1 | |
| Insertion | Chr3 | 21290123 | 21290123 | 1 | |
| Insertion | Chr5 | 20921055 | 20921056 | 2 | |
| Insertion | Chr7 | 21699879 | 21699900 | 22 | |
| Insertion | Chr7 | 28425797 | 28425797 | 1 | |
| Insertion | Chr8 | 14440936 | 14440936 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6701562 | 7084111 | 2 |
| Inversion | Chr5 | 6701574 | 7084132 | 2 |
No Translocation
