Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1540-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1540-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 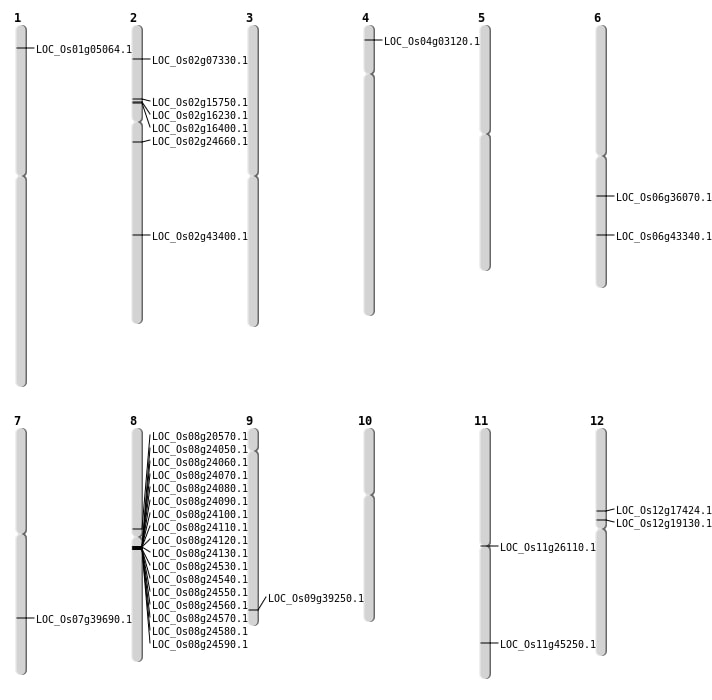
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 37110353 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 41132865 | G-T | Homo | INTERGENIC | |
| SBS | Chr1 | 8151655 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 12853748 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 14940458 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g26110 |
| SBS | Chr11 | 21050437 | A-G | Homo | INTERGENIC | |
| SBS | Chr11 | 21417941 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 26671416 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 27388322 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g45250 |
| SBS | Chr11 | 9350055 | G-T | HET | INTRON | |
| SBS | Chr12 | 10743748 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 11108140 | A-T | HET | STOP_GAINED | LOC_Os12g19130 |
| SBS | Chr12 | 21851126 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25454977 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 29084621 | T-C | Homo | INTRON | |
| SBS | Chr2 | 29486601 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9346930 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16400 |
| SBS | Chr3 | 13559989 | T-A | Homo | INTERGENIC | |
| SBS | Chr3 | 24508700 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 1299323 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03120 |
| SBS | Chr4 | 268987 | T-G | Homo | INTERGENIC | |
| SBS | Chr4 | 27287594 | T-A | HET | INTRON | |
| SBS | Chr4 | 28299450 | C-A | HET | INTRON | |
| SBS | Chr5 | 12827777 | A-T | Homo | INTRON | |
| SBS | Chr5 | 1358777 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15219045 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24618489 | G-A | HET | INTRON | |
| SBS | Chr5 | 27002677 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 26052870 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g43340 |
| SBS | Chr6 | 3785560 | G-A | HET | INTRON | |
| SBS | Chr6 | 3785561 | C-A | HET | INTRON | |
| SBS | Chr7 | 23876059 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 23952032 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 13091286 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 13108028 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 13108029 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 14495079 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 19553328 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 22561780 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 2961197 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 7424005 | A-G | HET | INTRON | |
| SBS | Chr9 | 22548434 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g39250 |
| SBS | Chr9 | 2990831 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 5294694 | T-A | HET | INTRON |
Deletions: 47
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1140651 | 1140652 | 1 | |
| Deletion | Chr2 | 3773884 | 3773899 | 15 | LOC_Os02g07330 |
| Deletion | Chr9 | 4014674 | 4014679 | 5 | |
| Deletion | Chr12 | 4764436 | 4764443 | 7 | |
| Deletion | Chr4 | 5038570 | 5038571 | 1 | |
| Deletion | Chr1 | 5357247 | 5357251 | 4 | |
| Deletion | Chr8 | 5807996 | 5807997 | 1 | |
| Deletion | Chr8 | 6711427 | 6711453 | 26 | |
| Deletion | Chr2 | 8876043 | 8876049 | 6 | LOC_Os02g15750 |
| Deletion | Chr2 | 9004995 | 9005008 | 13 | |
| Deletion | Chr2 | 9223591 | 9223602 | 11 | LOC_Os02g16230 |
| Deletion | Chr12 | 9992697 | 9992709 | 12 | LOC_Os12g17424 |
| Deletion | Chr4 | 10158058 | 10158064 | 6 | |
| Deletion | Chr11 | 10934850 | 10934866 | 16 | |
| Deletion | Chr1 | 11777722 | 11777736 | 14 | |
| Deletion | Chr3 | 12094166 | 12094167 | 1 | |
| Deletion | Chr11 | 12766849 | 12766852 | 3 | |
| Deletion | Chr11 | 12835720 | 12835725 | 5 | |
| Deletion | Chr3 | 13348416 | 13348417 | 1 | |
| Deletion | Chr2 | 14305804 | 14305806 | 2 | LOC_Os02g24660 |
| Deletion | Chr8 | 14544001 | 14580000 | 36000 | 9 |
| Deletion | Chr2 | 14551685 | 14551687 | 2 | |
| Deletion | Chr4 | 14633790 | 14633791 | 1 | |
| Deletion | Chr8 | 14811001 | 14875000 | 64000 | 7 |
| Deletion | Chr7 | 16124930 | 16124931 | 1 | |
| Deletion | Chr3 | 16355128 | 16355148 | 20 | |
| Deletion | Chr2 | 16589453 | 16589454 | 1 | |
| Deletion | Chr2 | 16761704 | 16761720 | 16 | |
| Deletion | Chr11 | 18637846 | 18637847 | 1 | |
| Deletion | Chr6 | 21091100 | 21091101 | 1 | LOC_Os06g36070 |
| Deletion | Chr8 | 22143805 | 22143818 | 13 | |
| Deletion | Chr8 | 22606823 | 22606859 | 36 | |
| Deletion | Chr8 | 22985603 | 22985613 | 10 | |
| Deletion | Chr4 | 23550883 | 23550884 | 1 | |
| Deletion | Chr7 | 23785377 | 23785400 | 23 | LOC_Os07g39690 |
| Deletion | Chr6 | 23952746 | 23952753 | 7 | |
| Deletion | Chr12 | 24513920 | 24513921 | 1 | |
| Deletion | Chr8 | 24547563 | 24547571 | 8 | |
| Deletion | Chr6 | 25488349 | 25488350 | 1 | |
| Deletion | Chr3 | 25492868 | 25492886 | 18 | |
| Deletion | Chr2 | 25544977 | 25544978 | 1 | |
| Deletion | Chr5 | 26272127 | 26272128 | 1 | |
| Deletion | Chr12 | 26607112 | 26607113 | 1 | |
| Deletion | Chr3 | 27485722 | 27485724 | 2 | |
| Deletion | Chr1 | 30816716 | 30817736 | 1020 | |
| Deletion | Chr1 | 31626963 | 31626964 | 1 | |
| Deletion | Chr3 | 32216685 | 32216686 | 1 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12362031 | 12362034 | 4 | |
| Insertion | Chr1 | 2377850 | 2377855 | 6 | LOC_Os01g05064 |
| Insertion | Chr10 | 7138490 | 7138490 | 1 | |
| Insertion | Chr12 | 24258197 | 24258197 | 1 | |
| Insertion | Chr12 | 5315490 | 5315490 | 1 | |
| Insertion | Chr12 | 6046886 | 6046886 | 1 | |
| Insertion | Chr2 | 25564944 | 25564965 | 22 | |
| Insertion | Chr2 | 28165371 | 28165384 | 14 | |
| Insertion | Chr2 | 28613584 | 28613635 | 52 | |
| Insertion | Chr3 | 7404429 | 7404448 | 20 | |
| Insertion | Chr4 | 12363255 | 12363258 | 4 | |
| Insertion | Chr4 | 2242568 | 2242568 | 1 | |
| Insertion | Chr5 | 11182632 | 11182633 | 2 | |
| Insertion | Chr6 | 22075505 | 22075507 | 3 | |
| Insertion | Chr8 | 20085411 | 20085412 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2982898 | 3181507 | |
| Inversion | Chr3 | 3181515 | 3280355 | |
| Inversion | Chr8 | 12353779 | 12999858 | LOC_Os08g20570 |
| Inversion | Chr8 | 12355081 | 12999888 | LOC_Os08g20570 |
| Inversion | Chr5 | 22515160 | 27300379 | |
| Inversion | Chr5 | 22515186 | 27300419 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 2982902 | Chr2 | 26193205 | LOC_Os02g43400 |
| Translocation | Chr3 | 3280348 | Chr2 | 26193894 | LOC_Os02g43400 |
| Translocation | Chr11 | 14321747 | Chr8 | 12353778 | LOC_Os08g20570 |
| Translocation | Chr11 | 14321786 | Chr8 | 12355101 | LOC_Os08g20570 |
| Translocation | Chr12 | 17386184 | Chr8 | 17758594 |
