Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1556-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1556-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 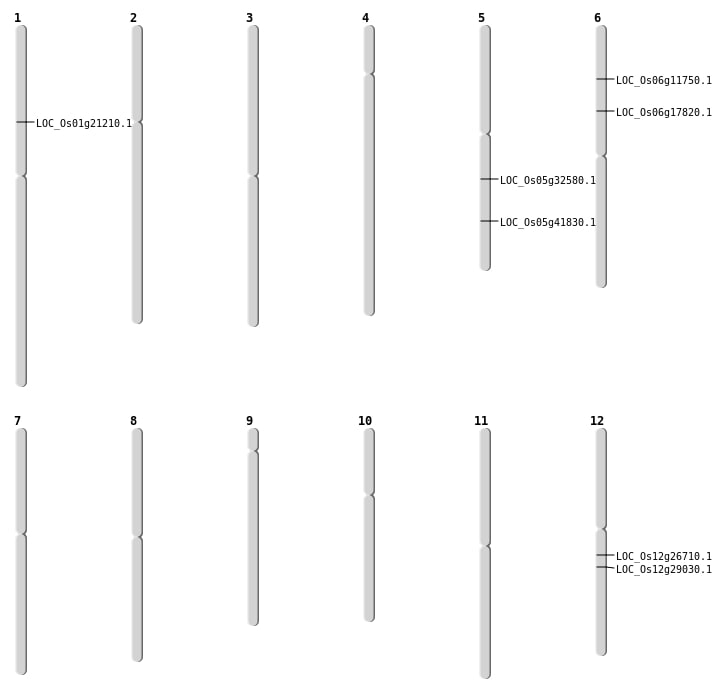
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16054754 | G-A | Homo | UTR_3_PRIME | |
| SBS | Chr1 | 16902509 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22602931 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 28962461 | C-T | Homo | INTERGENIC | |
| SBS | Chr10 | 2296033 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 21092149 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10567518 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 13912591 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 13912592 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 14027300 | T-A | HET | INTRON | |
| SBS | Chr12 | 17190377 | G-A | HET | STOP_GAINED | LOC_Os12g29030 |
| SBS | Chr12 | 19967854 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12888972 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 16147394 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 12169676 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 18699629 | G-C | Homo | INTERGENIC | |
| SBS | Chr3 | 2989542 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 2989543 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 2989544 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 12711656 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 13090814 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 16339023 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19371078 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 22920551 | G-A | HET | INTRON | |
| SBS | Chr4 | 3825208 | T-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr5 | 3579783 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 2817414 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 17853750 | C-T | Homo | INTRON | |
| SBS | Chr7 | 2396868 | A-G | Homo | INTERGENIC | |
| SBS | Chr8 | 15307845 | G-A | Homo | INTRON | |
| SBS | Chr8 | 18347535 | C-A | Homo | INTRON | |
| SBS | Chr8 | 19010561 | G-C | Homo | INTERGENIC | |
| SBS | Chr8 | 3943340 | C-T | HET | INTRON | |
| SBS | Chr9 | 670378 | A-G | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1698119 | 1698142 | 23 | |
| Deletion | Chr3 | 1942987 | 1942988 | 1 | |
| Deletion | Chr5 | 2154092 | 2154093 | 1 | |
| Deletion | Chr5 | 4841142 | 4841143 | 1 | |
| Deletion | Chr7 | 5227722 | 5227733 | 11 | |
| Deletion | Chr9 | 5571354 | 5571355 | 1 | |
| Deletion | Chr1 | 6260669 | 6260674 | 5 | |
| Deletion | Chr7 | 8067604 | 8067613 | 9 | |
| Deletion | Chr4 | 13211524 | 13211525 | 1 | |
| Deletion | Chr2 | 13308213 | 13308218 | 5 | |
| Deletion | Chr4 | 13345833 | 13345837 | 4 | |
| Deletion | Chr5 | 13760205 | 13760217 | 12 | |
| Deletion | Chr2 | 14526017 | 14526018 | 1 | |
| Deletion | Chr3 | 18146623 | 18146629 | 6 | |
| Deletion | Chr9 | 18399984 | 18399985 | 1 | |
| Deletion | Chr4 | 22739525 | 22739526 | 1 | |
| Deletion | Chr5 | 24498360 | 24498361 | 1 | LOC_Os05g41830 |
| Deletion | Chr1 | 26013566 | 26014227 | 661 | |
| Deletion | Chr2 | 26577556 | 26577558 | 2 | |
| Deletion | Chr7 | 26843150 | 26843162 | 12 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr7 | 28335216 | 28335218 | 2 | |
| Deletion | Chr6 | 29190528 | 29190532 | 4 | |
| Deletion | Chr5 | 29560011 | 29560014 | 3 | |
| Deletion | Chr2 | 30816847 | 30816851 | 4 | |
| Deletion | Chr2 | 31627041 | 31627045 | 4 | |
| Deletion | Chr2 | 34626689 | 34626690 | 1 | |
| Deletion | Chr1 | 37545585 | 37545586 | 1 |
Insertions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 3246058 | 3246058 | 1 | |
| Insertion | Chr12 | 20179144 | 20179146 | 3 | |
| Insertion | Chr12 | 20273415 | 20273416 | 2 | |
| Insertion | Chr12 | 8086645 | 8086645 | 1 | |
| Insertion | Chr2 | 17000809 | 17000855 | 47 | |
| Insertion | Chr3 | 23125839 | 23125844 | 6 | |
| Insertion | Chr3 | 25528256 | 25528276 | 21 | |
| Insertion | Chr3 | 25768971 | 25768971 | 1 | |
| Insertion | Chr3 | 28982499 | 28982500 | 2 | |
| Insertion | Chr4 | 28949468 | 28949468 | 1 | |
| Insertion | Chr4 | 35341328 | 35341328 | 1 | |
| Insertion | Chr5 | 16875749 | 16875749 | 1 | |
| Insertion | Chr5 | 19071866 | 19071868 | 3 | LOC_Os05g32580 |
| Insertion | Chr5 | 27863457 | 27863460 | 4 | |
| Insertion | Chr6 | 10332334 | 10332334 | 1 | LOC_Os06g17820 |
| Insertion | Chr7 | 24240074 | 24240104 | 31 | |
| Insertion | Chr9 | 4100788 | 4100788 | 1 | |
| Insertion | Chr9 | 8846950 | 8846991 | 42 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 5500194 | 5507540 | |
| Inversion | Chr6 | 6182767 | 6228647 | LOC_Os06g11750 |
| Inversion | Chr12 | 13160603 | 15654255 | LOC_Os12g26710 |
| Inversion | Chr12 | 13160617 | 15654259 | LOC_Os12g26710 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 982477 | Chr1 | 11835703 | LOC_Os01g21210 |
| Translocation | Chr12 | 982485 | Chr1 | 11836706 | LOC_Os01g21210 |
