Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1559-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1559-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 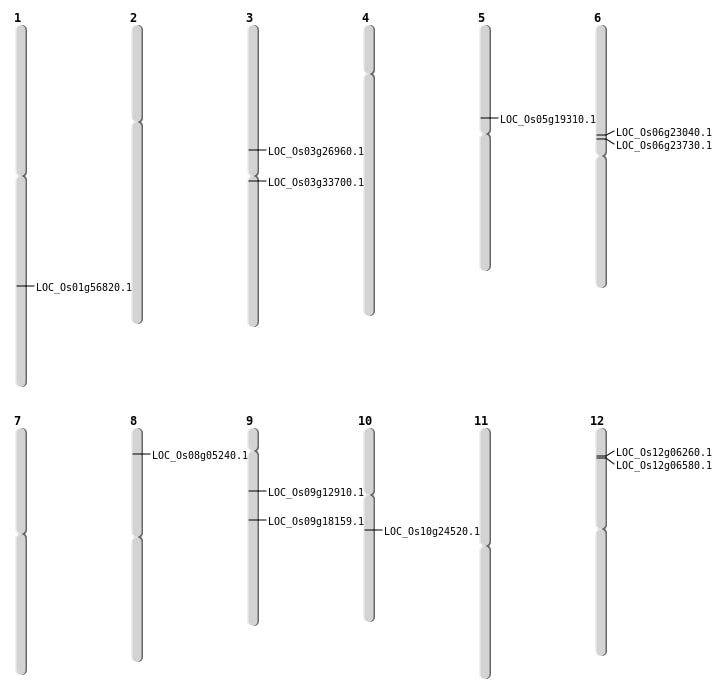
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10051794 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 13683846 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 16601881 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 17610435 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 17610446 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 24338155 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 32817984 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g56820 |
| SBS | Chr1 | 6378936 | C-T | Homo | INTRON | |
| SBS | Chr1 | 6378937 | A-C | Homo | INTRON | |
| SBS | Chr10 | 12587234 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24520 |
| SBS | Chr10 | 15468601 | C-G | Homo | INTERGENIC | |
| SBS | Chr10 | 26902 | G-A | Homo | INTERGENIC | |
| SBS | Chr10 | 3824825 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 9644226 | A-G | HET | INTRON | |
| SBS | Chr11 | 18944413 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17145481 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 1580074 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 26304188 | T-G | Homo | INTERGENIC | |
| SBS | Chr2 | 29830175 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr3 | 14127239 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 15413771 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g26960 |
| SBS | Chr3 | 19272508 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33700 |
| SBS | Chr3 | 20481567 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 29025283 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 23624265 | G-T | Homo | INTERGENIC | |
| SBS | Chr4 | 3964304 | A-C | Homo | INTERGENIC | |
| SBS | Chr5 | 11477980 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 10202955 | G-C | Homo | INTERGENIC | |
| SBS | Chr6 | 14026068 | C-A | Homo | INTERGENIC | |
| SBS | Chr6 | 4343114 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 938723 | A-G | Homo | INTRON | |
| SBS | Chr7 | 19044128 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 25649053 | C-A | Homo | UTR_5_PRIME | |
| SBS | Chr8 | 2725878 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g05240 |
| SBS | Chr8 | 8807498 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 8832951 | T-G | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 3503148 | 3503149 | 1 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr7 | 6864161 | 6864164 | 3 | |
| Deletion | Chr9 | 7614956 | 7614957 | 1 | |
| Deletion | Chr10 | 7668455 | 7668476 | 21 | |
| Deletion | Chr5 | 7929188 | 7929193 | 5 | |
| Deletion | Chr1 | 8744515 | 8744516 | 1 | |
| Deletion | Chr2 | 9418179 | 9418181 | 2 | |
| Deletion | Chr9 | 10471879 | 10472007 | 128 | |
| Deletion | Chr5 | 11225195 | 11225210 | 15 | LOC_Os05g19310 |
| Deletion | Chr11 | 11735399 | 11735404 | 5 | |
| Deletion | Chr2 | 12079171 | 12079172 | 1 | |
| Deletion | Chr3 | 13678381 | 13678390 | 9 | |
| Deletion | Chr1 | 14293372 | 14293390 | 18 | |
| Deletion | Chr6 | 14857211 | 14857212 | 1 | |
| Deletion | Chr11 | 16562250 | 16562254 | 4 | |
| Deletion | Chr12 | 17648776 | 17648790 | 14 | |
| Deletion | Chr10 | 18119507 | 18119512 | 5 | |
| Deletion | Chr4 | 23578503 | 23578505 | 2 | |
| Deletion | Chr6 | 23677324 | 23677325 | 1 | |
| Deletion | Chr12 | 23924680 | 23924682 | 2 | |
| Deletion | Chr6 | 24322010 | 24322028 | 18 | |
| Deletion | Chr11 | 24399163 | 24399168 | 5 | |
| Deletion | Chr5 | 25660375 | 25660377 | 2 | |
| Deletion | Chr1 | 38952881 | 38952895 | 14 |
Insertions: 11
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 2979479 | 3193048 | 2 |
| Inversion | Chr9 | 7435641 | 11121071 | |
| Inversion | Chr9 | 7439012 | 11132047 | 2 |
| Inversion | Chr6 | 13442360 | 13868415 | 2 |
No Translocation
