Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1561-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1561-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 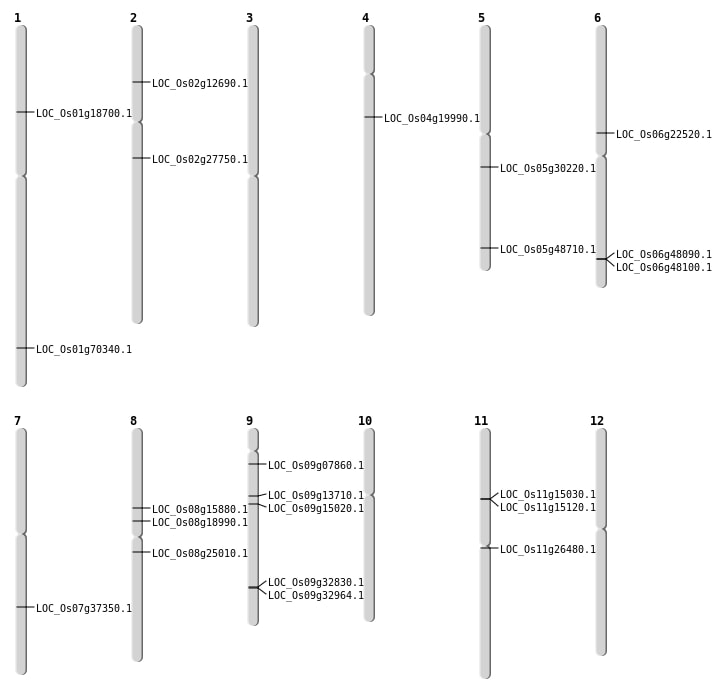
|
| Variant Information |
|---|
Single base substitutions: 60
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10546350 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g18700 |
| SBS | Chr1 | 13723501 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 15232696 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 15664982 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 19685526 | A-T | HET | INTRON | |
| SBS | Chr1 | 24292437 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 39667120 | G-A | HET | INTRON | |
| SBS | Chr10 | 11061856 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 15038340 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 9351973 | G-A | HET | INTRON | |
| SBS | Chr11 | 10844075 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 13256182 | G-A | HET | INTRON | |
| SBS | Chr11 | 13928659 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15175691 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26480 |
| SBS | Chr11 | 16798793 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 20177120 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 25434957 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9047887 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 18801542 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 18801543 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 26357669 | A-T | Homo | INTERGENIC | |
| SBS | Chr12 | 5665446 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 6919730 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8032818 | T-C | HET | INTRON | |
| SBS | Chr2 | 2791140 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 29921110 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 3598467 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 19366749 | A-G | HET | INTRON | |
| SBS | Chr3 | 20832214 | C-T | Homo | INTRON | |
| SBS | Chr3 | 23248550 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 30157618 | C-T | Homo | INTRON | |
| SBS | Chr3 | 4512859 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 7129959 | C-T | Homo | INTRON | |
| SBS | Chr4 | 10630720 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 11145129 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19990 |
| SBS | Chr4 | 16965101 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20270409 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2902454 | C-T | HET | INTRON | |
| SBS | Chr5 | 12804950 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 14170159 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17516657 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g30220 |
| SBS | Chr5 | 27210441 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 27923625 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g48710 |
| SBS | Chr5 | 5556666 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 17973048 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 24284966 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 4767770 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 12403177 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 13813346 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 24826593 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 25703346 | A-T | HET | INTRON | |
| SBS | Chr8 | 27540871 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 9670621 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g15880 |
| SBS | Chr9 | 10077839 | C-A | HET | INTRON | |
| SBS | Chr9 | 19674789 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19674790 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g32964 |
| SBS | Chr9 | 22410881 | G-A | HET | INTRON | |
| SBS | Chr9 | 3997712 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
| SBS | Chr9 | 5235311 | T-C | HET | INTRON | |
| SBS | Chr9 | 697206 | C-T | HET | INTRON |
Deletions: 47
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 170870 | 170873 | 3 | |
| Deletion | Chr10 | 609301 | 609302 | 1 | |
| Deletion | Chr7 | 3855484 | 3855486 | 2 | |
| Deletion | Chr4 | 5066942 | 5066943 | 1 | |
| Deletion | Chr2 | 6638812 | 6638820 | 8 | LOC_Os02g12690 |
| Deletion | Chr10 | 7770267 | 7770268 | 1 | |
| Deletion | Chr9 | 8023478 | 8023481 | 3 | LOC_Os09g13710 |
| Deletion | Chr1 | 8219688 | 8219690 | 2 | |
| Deletion | Chr9 | 8610790 | 8610791 | 1 | |
| Deletion | Chr4 | 8774490 | 8774494 | 4 | |
| Deletion | Chr9 | 9055287 | 9055288 | 1 | LOC_Os09g15020 |
| Deletion | Chr7 | 9728966 | 9728969 | 3 | |
| Deletion | Chr8 | 9912385 | 9912387 | 2 | |
| Deletion | Chr1 | 10242633 | 10242634 | 1 | |
| Deletion | Chr9 | 10816466 | 10816467 | 1 | |
| Deletion | Chr4 | 11128621 | 11128622 | 1 | |
| Deletion | Chr6 | 11183396 | 11183418 | 22 | |
| Deletion | Chr8 | 11336106 | 11336107 | 1 | LOC_Os08g18990 |
| Deletion | Chr7 | 11354958 | 11354960 | 2 | |
| Deletion | Chr6 | 13029627 | 13029628 | 1 | |
| Deletion | Chr6 | 13086576 | 13086584 | 8 | LOC_Os06g22520 |
| Deletion | Chr8 | 13898483 | 13898484 | 1 | |
| Deletion | Chr8 | 14441033 | 14441034 | 1 | |
| Deletion | Chr11 | 14806545 | 14806553 | 8 | |
| Deletion | Chr1 | 16290790 | 16290791 | 1 | |
| Deletion | Chr2 | 16436186 | 16436187 | 1 | LOC_Os02g27750 |
| Deletion | Chr4 | 16812853 | 16812854 | 1 | |
| Deletion | Chr4 | 17016600 | 17016612 | 12 | |
| Deletion | Chr2 | 18802620 | 18802621 | 1 | |
| Deletion | Chr12 | 18897960 | 18897963 | 3 | |
| Deletion | Chr7 | 19245128 | 19245131 | 3 | |
| Deletion | Chr9 | 19575681 | 19581250 | 5569 | LOC_Os09g32830 |
| Deletion | Chr12 | 19761757 | 19761768 | 11 | |
| Deletion | Chr6 | 20382925 | 20382928 | 3 | |
| Deletion | Chr6 | 20496905 | 20496906 | 1 | |
| Deletion | Chr7 | 22370967 | 22370974 | 7 | LOC_Os07g37350 |
| Deletion | Chr4 | 23435488 | 23435495 | 7 | |
| Deletion | Chr3 | 24070052 | 24070059 | 7 | |
| Deletion | Chr6 | 24604853 | 24604854 | 1 | |
| Deletion | Chr6 | 24658443 | 24658464 | 21 | |
| Deletion | Chr4 | 26335759 | 26335763 | 4 | |
| Deletion | Chr5 | 27791966 | 27791967 | 1 | |
| Deletion | Chr6 | 29095453 | 29107115 | 11662 | 2 |
| Deletion | Chr2 | 31215776 | 31215777 | 1 | |
| Deletion | Chr3 | 31637977 | 31637983 | 6 | |
| Deletion | Chr1 | 39533490 | 39533514 | 24 | |
| Deletion | Chr1 | 40516455 | 40516456 | 1 |
Insertions: 12
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 8457811 | 8476601 | LOC_Os11g15030 |
| Inversion | Chr11 | 8476603 | 8513902 | LOC_Os11g15120 |
| Inversion | Chr8 | 15166096 | 15237330 | LOC_Os08g25010 |
| Inversion | Chr8 | 15237319 | 15320761 | |
| Inversion | Chr1 | 40371445 | 40748258 | LOC_Os01g70340 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8457815 | Chr4 | 34434699 | LOC_Os11g15030 |
| Translocation | Chr11 | 8458705 | Chr4 | 34435452 | LOC_Os11g15030 |
| Translocation | Chr6 | 15747380 | Chr4 | 19596842 |
