Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1563-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1563-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 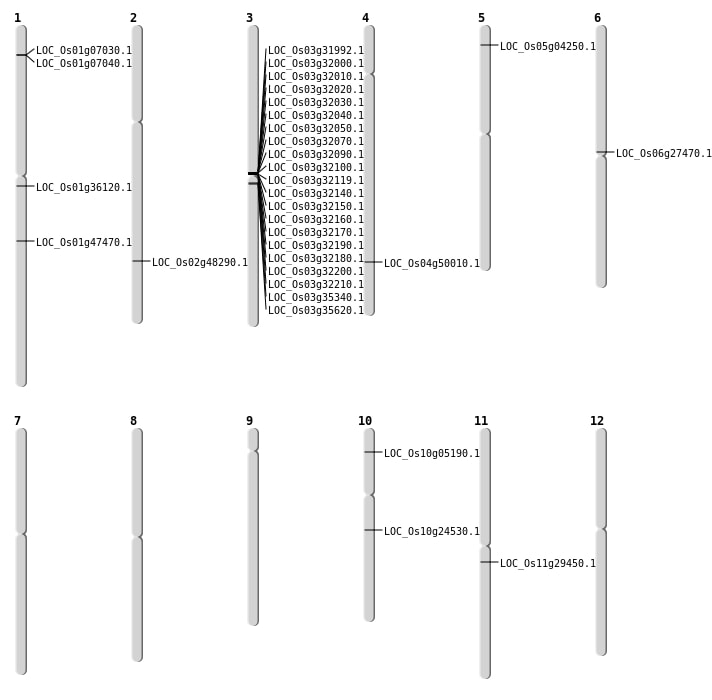
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17951280 | A-T | Homo | INTRON | |
| SBS | Chr1 | 28124056 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34079544 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 36861452 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 9307313 | T-G | Homo | SYNONYMOUS_CODING | |
| SBS | Chr10 | 12117486 | G-A | Homo | INTERGENIC | |
| SBS | Chr10 | 12590164 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g24530 |
| SBS | Chr10 | 9198301 | A-T | Homo | INTERGENIC | |
| SBS | Chr11 | 15168301 | A-G | Homo | INTERGENIC | |
| SBS | Chr11 | 17093579 | G-A | Homo | STOP_GAINED | LOC_Os11g29450 |
| SBS | Chr11 | 25515776 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 27143135 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 2137491 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 2137493 | T-A | Homo | INTERGENIC | |
| SBS | Chr2 | 29551509 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g48290 |
| SBS | Chr2 | 29551510 | C-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27054328 | T-A | Homo | INTERGENIC | |
| SBS | Chr4 | 12799540 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 17081042 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 1936660 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g04250 |
| SBS | Chr5 | 23726251 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 15553460 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g27470 |
| SBS | Chr6 | 23042672 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8712959 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 10483236 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 13271855 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 10244432 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 26568155 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 7744849 | G-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2040349 | 2040353 | 4 | |
| Deletion | Chr1 | 3312874 | 3315206 | 2332 | 2 |
| Deletion | Chr6 | 4214009 | 4214012 | 3 | |
| Deletion | Chr2 | 4647994 | 4647995 | 1 | |
| Deletion | Chr10 | 6740454 | 6740455 | 1 | |
| Deletion | Chr3 | 7333578 | 7333589 | 11 | |
| Deletion | Chr6 | 8927647 | 8927657 | 10 | |
| Deletion | Chr1 | 13172887 | 13172888 | 1 | |
| Deletion | Chr3 | 14556294 | 14556329 | 35 | |
| Deletion | Chr3 | 18215405 | 18215414 | 9 | |
| Deletion | Chr3 | 18300001 | 18428000 | 128000 | 19 |
| Deletion | Chr7 | 18708336 | 18708343 | 7 | |
| Deletion | Chr12 | 19915787 | 19915799 | 12 | |
| Deletion | Chr1 | 19983252 | 19983253 | 1 | LOC_Os01g36120 |
| Deletion | Chr8 | 20191517 | 20191519 | 2 | |
| Deletion | Chr9 | 20231379 | 20231380 | 1 | |
| Deletion | Chr8 | 25434676 | 25434678 | 2 | |
| Deletion | Chr3 | 26766752 | 26766761 | 9 | |
| Deletion | Chr5 | 27762798 | 27762806 | 8 | |
| Deletion | Chr1 | 28471704 | 28471714 | 10 | |
| Deletion | Chr1 | 29168337 | 29168358 | 21 | |
| Deletion | Chr4 | 29823676 | 29823693 | 17 | LOC_Os04g50010 |
| Deletion | Chr4 | 34100214 | 34100220 | 6 | |
| Deletion | Chr1 | 35977553 | 35977555 | 2 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 27134019 | 27134020 | 2 | LOC_Os01g47470 |
| Insertion | Chr1 | 35770112 | 35770113 | 2 | |
| Insertion | Chr10 | 18605262 | 18605321 | 60 | |
| Insertion | Chr10 | 2543903 | 2543935 | 33 | LOC_Os10g05190 |
| Insertion | Chr12 | 17141968 | 17141969 | 2 | |
| Insertion | Chr2 | 1506254 | 1506255 | 2 | |
| Insertion | Chr2 | 29632464 | 29632464 | 1 | |
| Insertion | Chr3 | 3505290 | 3505290 | 1 | |
| Insertion | Chr3 | 9783060 | 9783060 | 1 | |
| Insertion | Chr4 | 18853573 | 18853616 | 44 | |
| Insertion | Chr4 | 2305618 | 2305618 | 1 | |
| Insertion | Chr6 | 15988354 | 15988355 | 2 | |
| Insertion | Chr6 | 8981674 | 8981675 | 2 | |
| Insertion | Chr8 | 14445514 | 14445515 | 2 | |
| Insertion | Chr8 | 15137422 | 15137423 | 2 | |
| Insertion | Chr9 | 22474988 | 22475044 | 57 | |
| Insertion | Chr9 | 4781773 | 4781774 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2230406 | 2395465 | |
| Inversion | Chr3 | 19594721 | 19737436 | 2 |
| Inversion | Chr3 | 19597153 | 19922450 | LOC_Os03g35340 |
| Inversion | Chr12 | 24848070 | 24917872 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6287691 | Chr3 | 19597146 | LOC_Os03g35340 |
| Translocation | Chr5 | 6287695 | Chr3 | 19594748 | LOC_Os03g35340 |
