Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1565-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1565-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 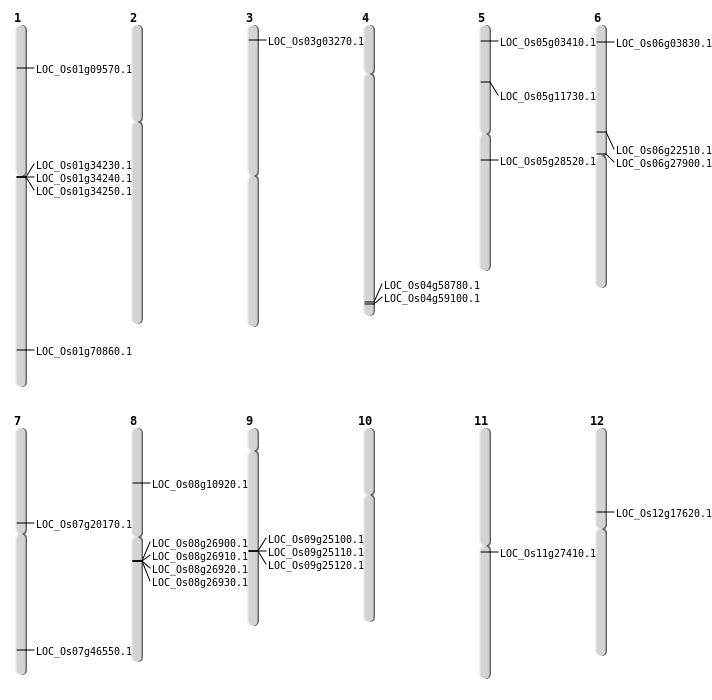
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31300977 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 9134165 | C-T | HET | INTRON | |
| SBS | Chr10 | 13842581 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 10581486 | G-T | HET | INTRON | |
| SBS | Chr12 | 10092483 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17620 |
| SBS | Chr12 | 14409151 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr2 | 13871482 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 13965549 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 15211763 | A-G | HET | INTRON | |
| SBS | Chr3 | 1403813 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 1403814 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g03270 |
| SBS | Chr3 | 24547225 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 13643676 | C-T | HET | INTRON | |
| SBS | Chr4 | 29559382 | T-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 8623823 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1056479 | A-C | Homo | INTERGENIC | |
| SBS | Chr5 | 17728207 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 7922188 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 9361426 | A-T | HET | INTRON | |
| SBS | Chr6 | 10714019 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13421983 | C-T | HET | INTRON | |
| SBS | Chr6 | 1539342 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g03830 |
| SBS | Chr6 | 15835133 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g27900 |
| SBS | Chr6 | 26326080 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 27083846 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 7957747 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11644610 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g20170 |
| SBS | Chr7 | 18220433 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 26079906 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 27800586 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46550 |
| SBS | Chr7 | 7296632 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17862621 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 6618116 | G-A | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 325308 | 325309 | 1 | |
| Deletion | Chr5 | 1418980 | 1418994 | 14 | LOC_Os05g03410 |
| Deletion | Chr4 | 3131103 | 3131118 | 15 | |
| Deletion | Chr5 | 3202191 | 3202217 | 26 | |
| Deletion | Chr5 | 3223267 | 3223268 | 1 | |
| Deletion | Chr8 | 6616602 | 6616621 | 19 | |
| Deletion | Chr5 | 8451850 | 8451851 | 1 | |
| Deletion | Chr9 | 9796427 | 9796430 | 3 | |
| Deletion | Chr6 | 10858551 | 10858555 | 4 | |
| Deletion | Chr6 | 13081288 | 13082914 | 1626 | LOC_Os06g22510 |
| Deletion | Chr7 | 13733594 | 13733595 | 1 | |
| Deletion | Chr9 | 15022001 | 15055000 | 33000 | 3 |
| Deletion | Chr11 | 15339862 | 15339864 | 2 | |
| Deletion | Chr11 | 15774660 | 15774663 | 3 | LOC_Os11g27410 |
| Deletion | Chr7 | 16268550 | 16268578 | 28 | |
| Deletion | Chr8 | 16403001 | 16434000 | 31000 | 4 |
| Deletion | Chr5 | 16704311 | 16704319 | 8 | LOC_Os05g28520 |
| Deletion | Chr1 | 18874130 | 18880574 | 6444 | 3 |
| Deletion | Chr6 | 19598851 | 19598856 | 5 | |
| Deletion | Chr9 | 21604482 | 21604495 | 13 | |
| Deletion | Chr4 | 22571352 | 22571358 | 6 | |
| Deletion | Chr7 | 24084870 | 24084883 | 13 | |
| Deletion | Chr7 | 24161355 | 24161359 | 4 | |
| Deletion | Chr6 | 24778654 | 24778664 | 10 | |
| Deletion | Chr5 | 25149287 | 25149303 | 16 | |
| Deletion | Chr1 | 27025804 | 27026182 | 378 | |
| Deletion | Chr2 | 30406480 | 30406482 | 2 | |
| Deletion | Chr2 | 34769003 | 34769012 | 9 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 38799864 | 38799865 | 2 | |
| Insertion | Chr1 | 4909131 | 4909134 | 4 | LOC_Os01g09570 |
| Insertion | Chr1 | 5284523 | 5284524 | 2 | |
| Insertion | Chr10 | 19118966 | 19118966 | 1 | |
| Insertion | Chr11 | 22130900 | 22130929 | 30 | |
| Insertion | Chr11 | 22790210 | 22790211 | 2 | |
| Insertion | Chr12 | 3653299 | 3653302 | 4 | |
| Insertion | Chr2 | 15876877 | 15876878 | 2 | |
| Insertion | Chr3 | 25670560 | 25670560 | 1 | |
| Insertion | Chr3 | 35283026 | 35283026 | 1 | |
| Insertion | Chr5 | 14802562 | 14802562 | 1 | |
| Insertion | Chr7 | 14601802 | 14601802 | 1 | |
| Insertion | Chr7 | 15855178 | 15855183 | 6 | |
| Insertion | Chr7 | 208063 | 208064 | 2 | |
| Insertion | Chr7 | 27412774 | 27412774 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 5256277 | 6432462 | LOC_Os08g10920 |
| Inversion | Chr5 | 6658731 | 7994713 | LOC_Os05g11730 |
| Inversion | Chr4 | 34962084 | 35161616 | 2 |
| Inversion | Chr1 | 40896618 | 41008621 | LOC_Os01g70860 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 19649004 | Chr1 | 18880597 | LOC_Os01g34250 |
