Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1572-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1572-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 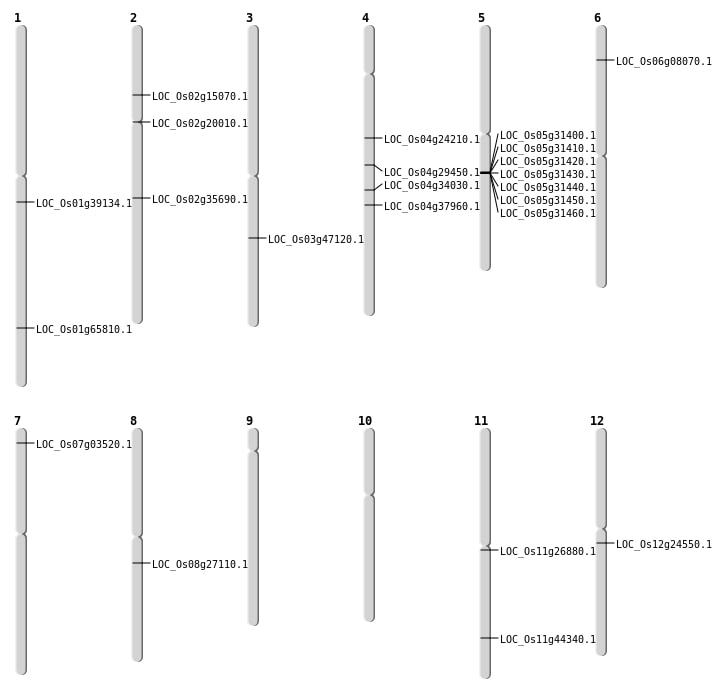
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17317010 | C-G | HET | INTRON | |
| SBS | Chr1 | 22023748 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39134 |
| SBS | Chr1 | 23876354 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 34057660 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 38220280 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g65810 |
| SBS | Chr10 | 15131277 | A-C | Homo | INTERGENIC | |
| SBS | Chr10 | 8966168 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 965356 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 10378174 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 23038297 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 316664 | C-T | Homo | INTERGENIC | |
| SBS | Chr11 | 7584810 | C-G | Homo | UTR_3_PRIME | |
| SBS | Chr11 | 8201250 | T-A | Homo | INTRON | |
| SBS | Chr12 | 14030721 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g24550 |
| SBS | Chr12 | 15304766 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 25858661 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 20530797 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 21450013 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g35690 |
| SBS | Chr2 | 23844830 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 3756077 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 8384036 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 8402922 | G-A | Homo | STOP_GAINED | LOC_Os02g15070 |
| SBS | Chr3 | 14735242 | G-A | HET | INTRON | |
| SBS | Chr3 | 14735244 | G-A | HET | INTRON | |
| SBS | Chr3 | 25218515 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 2702327 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10376462 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10635111 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 13856772 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24210 |
| SBS | Chr4 | 15051148 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2327588 | G-C | HET | INTRON | |
| SBS | Chr5 | 29652448 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 4602640 | G-T | HET | INTRON | |
| SBS | Chr5 | 482251 | A-T | Homo | INTERGENIC | |
| SBS | Chr5 | 929372 | T-G | Homo | INTRON | |
| SBS | Chr7 | 5689086 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 16567981 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27110 |
| SBS | Chr9 | 14737212 | T-C | Homo | INTERGENIC | |
| SBS | Chr9 | 18228192 | C-T | HET | INTRON |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 339564 | 339565 | 1 | |
| Deletion | Chr11 | 1052674 | 1052675 | 1 | |
| Deletion | Chr7 | 1405933 | 1405934 | 1 | LOC_Os07g03520 |
| Deletion | Chr3 | 2459293 | 2459300 | 7 | |
| Deletion | Chr3 | 2629229 | 2629235 | 6 | |
| Deletion | Chr6 | 3903740 | 3903741 | 1 | LOC_Os06g08070 |
| Deletion | Chr11 | 3949172 | 3949173 | 1 | |
| Deletion | Chr10 | 4011324 | 4011328 | 4 | |
| Deletion | Chr10 | 8419283 | 8419303 | 20 | |
| Deletion | Chr2 | 10344164 | 10344173 | 9 | |
| Deletion | Chr2 | 11781594 | 11781596 | 2 | LOC_Os02g20010 |
| Deletion | Chr9 | 11963444 | 11963445 | 1 | |
| Deletion | Chr8 | 15056860 | 15056862 | 2 | |
| Deletion | Chr11 | 15445916 | 15445918 | 2 | LOC_Os11g26880 |
| Deletion | Chr3 | 15930731 | 15930735 | 4 | |
| Deletion | Chr1 | 15980148 | 15980149 | 1 | |
| Deletion | Chr11 | 16278872 | 16278874 | 2 | |
| Deletion | Chr4 | 17509994 | 17509997 | 3 | LOC_Os04g29450 |
| Deletion | Chr1 | 18103534 | 18103538 | 4 | |
| Deletion | Chr5 | 18261001 | 18301000 | 40000 | 7 |
| Deletion | Chr1 | 22030801 | 22030820 | 19 | |
| Deletion | Chr4 | 22444168 | 22444177 | 9 | |
| Deletion | Chr1 | 24247804 | 24247823 | 19 | |
| Deletion | Chr4 | 26280502 | 26280511 | 9 | |
| Deletion | Chr6 | 26324330 | 26324332 | 2 | |
| Deletion | Chr3 | 26651621 | 26651623 | 2 | LOC_Os03g47120 |
| Deletion | Chr11 | 26802247 | 26802248 | 1 | LOC_Os11g44340 |
| Deletion | Chr5 | 27640846 | 27640847 | 1 | |
| Deletion | Chr5 | 27640847 | 27640848 | 1 | |
| Deletion | Chr3 | 28059235 | 28059245 | 10 | |
| Deletion | Chr2 | 31095427 | 31095429 | 2 | |
| Deletion | Chr3 | 35435131 | 35436273 | 1142 |
Insertions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 26424691 | 26424691 | 1 | |
| Insertion | Chr1 | 5763703 | 5763704 | 2 | |
| Insertion | Chr11 | 20878014 | 20878015 | 2 | |
| Insertion | Chr11 | 9521670 | 9521673 | 4 | |
| Insertion | Chr12 | 21983154 | 21983156 | 3 | |
| Insertion | Chr12 | 7670531 | 7670531 | 1 | |
| Insertion | Chr2 | 18460158 | 18460203 | 46 | |
| Insertion | Chr3 | 16351803 | 16351803 | 1 | |
| Insertion | Chr3 | 18983750 | 18983752 | 3 | |
| Insertion | Chr3 | 21887146 | 21887147 | 2 | |
| Insertion | Chr3 | 25164238 | 25164239 | 2 | |
| Insertion | Chr4 | 20610492 | 20610566 | 75 | LOC_Os04g34030 |
| Insertion | Chr4 | 22587228 | 22587228 | 1 | LOC_Os04g37960 |
| Insertion | Chr4 | 24275655 | 24275656 | 2 | |
| Insertion | Chr4 | 26317313 | 26317313 | 1 | |
| Insertion | Chr5 | 19846466 | 19846466 | 1 | |
| Insertion | Chr5 | 27226910 | 27226980 | 71 | |
| Insertion | Chr6 | 18938673 | 18938673 | 1 | |
| Insertion | Chr6 | 448491 | 448491 | 1 | |
| Insertion | Chr7 | 12237763 | 12237763 | 1 | |
| Insertion | Chr7 | 23315021 | 23315021 | 1 | |
| Insertion | Chr9 | 14737227 | 14737227 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 20373422 | 20528420 | |
| Inversion | Chr3 | 26668444 | 26909289 | |
| Inversion | Chr3 | 26668456 | 26909293 |
No Translocation
