Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1602-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1602-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 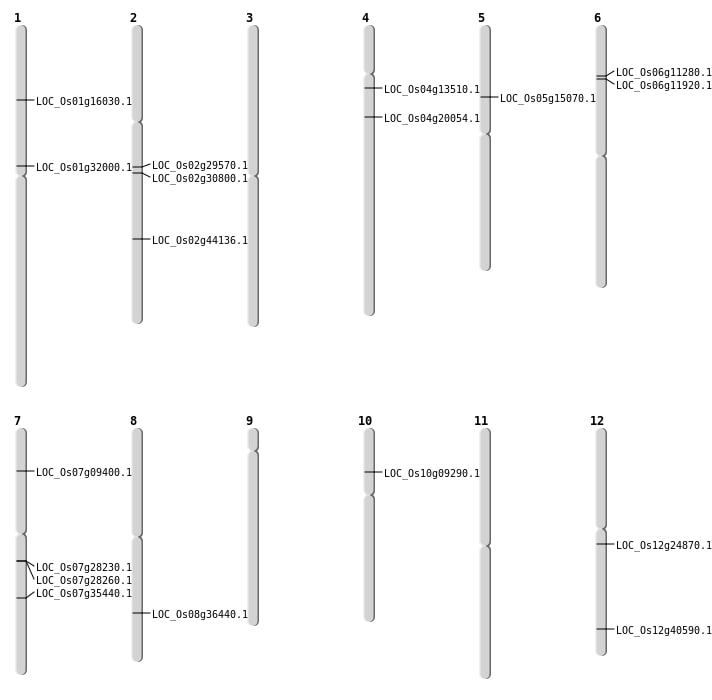
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18845021 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 24733844 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26482514 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 20860785 | G-T | Homo | INTERGENIC | |
| SBS | Chr10 | 5617307 | A-G | Homo | INTRON | |
| SBS | Chr10 | 7074441 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9894834 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 18576864 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 14259175 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g24870 |
| SBS | Chr12 | 22453002 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 22455917 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 25116628 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g40590 |
| SBS | Chr2 | 34076018 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 25019319 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 15651639 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 21101749 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 235655 | C-T | HET | INTRON | |
| SBS | Chr6 | 5681640 | A-T | Homo | UTR_3_PRIME | |
| SBS | Chr6 | 5913473 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g11280 |
| SBS | Chr7 | 16776463 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 4959596 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 4959597 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g09400 |
| SBS | Chr7 | 4959598 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g09400 |
| SBS | Chr8 | 12372464 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 1785015 | C-T | HET | INTRON | |
| SBS | Chr8 | 19566291 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 2173213 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22997370 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g36440 |
| SBS | Chr8 | 22997371 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15703674 | G-A | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 158845 | 158873 | 28 | |
| Deletion | Chr9 | 679459 | 679481 | 22 | |
| Deletion | Chr5 | 2120575 | 2120626 | 51 | |
| Deletion | Chr12 | 4109996 | 4109997 | 1 | |
| Deletion | Chr12 | 4391342 | 4391345 | 3 | |
| Deletion | Chr6 | 6351935 | 6351951 | 16 | LOC_Os06g11920 |
| Deletion | Chr5 | 7582322 | 7582323 | 1 | |
| Deletion | Chr5 | 8561659 | 8561721 | 62 | LOC_Os05g15070 |
| Deletion | Chr9 | 8909530 | 8909534 | 4 | |
| Deletion | Chr1 | 9023509 | 9023511 | 2 | LOC_Os01g16030 |
| Deletion | Chr8 | 10015254 | 10015255 | 1 | |
| Deletion | Chr11 | 10188795 | 10188796 | 1 | |
| Deletion | Chr1 | 11692734 | 11692736 | 2 | |
| Deletion | Chr12 | 12120466 | 12120468 | 2 | |
| Deletion | Chr1 | 14795471 | 14795477 | 6 | |
| Deletion | Chr3 | 14861878 | 14861880 | 2 | |
| Deletion | Chr2 | 15356632 | 15356643 | 11 | |
| Deletion | Chr5 | 15568322 | 15568328 | 6 | |
| Deletion | Chr9 | 16282318 | 16282319 | 1 | |
| Deletion | Chr7 | 17210582 | 17210616 | 34 | |
| Deletion | Chr3 | 17795871 | 17797730 | 1859 | |
| Deletion | Chr2 | 18364040 | 18364052 | 12 | LOC_Os02g30800 |
| Deletion | Chr7 | 18427700 | 18427704 | 4 | |
| Deletion | Chr4 | 20012995 | 20012996 | 1 | |
| Deletion | Chr5 | 21256537 | 21256545 | 8 | |
| Deletion | Chr9 | 22245374 | 22245379 | 5 | |
| Deletion | Chr12 | 22400881 | 22400882 | 1 | |
| Deletion | Chr12 | 22937389 | 22937395 | 6 | |
| Deletion | Chr7 | 25655275 | 25655282 | 7 | |
| Deletion | Chr3 | 28918139 | 28918141 | 2 | |
| Deletion | Chr6 | 31137929 | 31137933 | 4 | |
| Deletion | Chr4 | 34357601 | 34357602 | 1 | |
| Deletion | Chr1 | 36834561 | 36834587 | 26 | |
| Deletion | Chr1 | 41758090 | 41758092 | 2 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 25200701 | 25200701 | 1 | |
| Insertion | Chr11 | 25904717 | 25904718 | 2 | |
| Insertion | Chr2 | 11916433 | 11916433 | 1 | |
| Insertion | Chr2 | 18371365 | 18371366 | 2 | |
| Insertion | Chr2 | 23639566 | 23639566 | 1 | |
| Insertion | Chr4 | 11173495 | 11173544 | 50 | LOC_Os04g20054 |
| Insertion | Chr4 | 6134887 | 6134958 | 72 | |
| Insertion | Chr5 | 29813952 | 29813952 | 1 | |
| Insertion | Chr7 | 2573181 | 2573211 | 31 | |
| Insertion | Chr9 | 8981681 | 8981683 | 3 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 7535958 | 7536123 | LOC_Os04g13510 |
| Inversion | Chr7 | 16487925 | 16521720 | 2 |
| Inversion | Chr2 | 26676506 | 26713034 | LOC_Os02g44136 |
| Inversion | Chr1 | 33543520 | 33543783 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3684651 | Chr9 | 7178701 | |
| Translocation | Chr11 | 12580749 | Chr8 | 22301638 | |
| Translocation | Chr2 | 17593767 | Chr1 | 17520933 | 2 |
| Translocation | Chr12 | 17843383 | Chr10 | 5024981 | LOC_Os10g09290 |
| Translocation | Chr7 | 21194732 | Chr3 | 17797725 | LOC_Os07g35440 |
| Translocation | Chr11 | 23699280 | Chr4 | 29551823 |
