Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1617-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1617-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 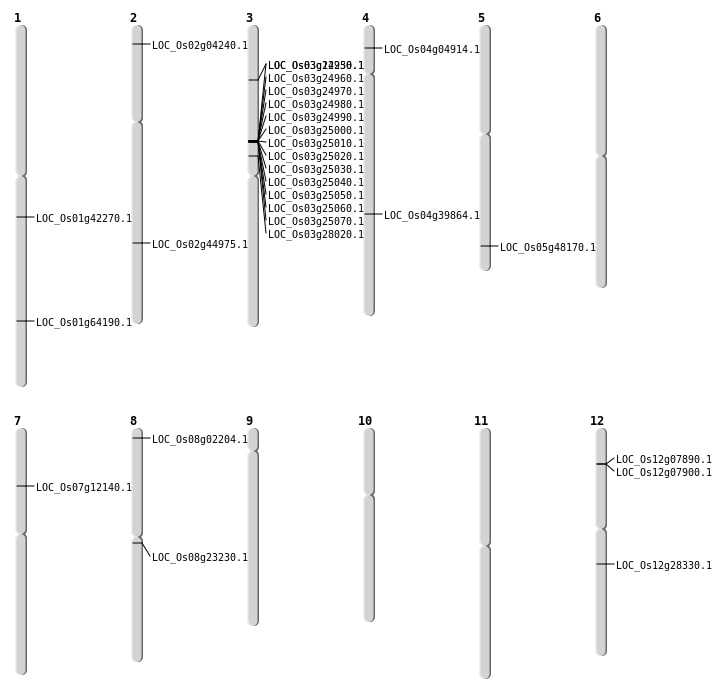
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26244124 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 29323403 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 34587426 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 41827538 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11492226 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 14318408 | C-G | Homo | INTERGENIC | |
| SBS | Chr11 | 23000969 | T-A | Homo | INTERGENIC | |
| SBS | Chr11 | 23000970 | T-A | Homo | INTERGENIC | |
| SBS | Chr12 | 16756382 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28330 |
| SBS | Chr12 | 21252615 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 4238928 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21090153 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 27197808 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 14336058 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 16105416 | C-T | Homo | STOP_GAINED | LOC_Os03g28020 |
| SBS | Chr3 | 18101646 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 2214559 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 24230687 | G-T | Homo | INTERGENIC | |
| SBS | Chr3 | 33683797 | C-G | HET | INTRON | |
| SBS | Chr4 | 10889671 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 2372159 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04914 |
| SBS | Chr4 | 23739206 | A-C | HET | INTRON | |
| SBS | Chr4 | 9014862 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 2699176 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 27610245 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48170 |
| SBS | Chr5 | 28116512 | C-A | Homo | INTERGENIC | |
| SBS | Chr5 | 3492547 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13653659 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 10574466 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19341461 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 24405229 | C-G | HET | INTRON | |
| SBS | Chr7 | 29495753 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 6795312 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g12140 |
| SBS | Chr8 | 14026605 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23230 |
| SBS | Chr8 | 5683258 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 11234113 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 1992724 | C-T | HET | INTRON | |
| SBS | Chr9 | 20848685 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 3455563 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 5000671 | C-T | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 750802 | 750804 | 2 | LOC_Os08g02204 |
| Deletion | Chr12 | 4000534 | 4007515 | 6981 | 2 |
| Deletion | Chr3 | 4421687 | 4421688 | 1 | |
| Deletion | Chr6 | 5806532 | 5806557 | 25 | |
| Deletion | Chr4 | 6460453 | 6460454 | 1 | |
| Deletion | Chr1 | 7486635 | 7486636 | 1 | |
| Deletion | Chr7 | 7797921 | 7797922 | 1 | |
| Deletion | Chr6 | 8338915 | 8338918 | 3 | |
| Deletion | Chr12 | 9640589 | 9640590 | 1 | |
| Deletion | Chr2 | 11913615 | 11913616 | 1 | |
| Deletion | Chr10 | 13123027 | 13123031 | 4 | |
| Deletion | Chr1 | 13673150 | 13673196 | 46 | |
| Deletion | Chr4 | 14006466 | 14006468 | 2 | |
| Deletion | Chr3 | 14226001 | 14316000 | 90000 | 12 |
| Deletion | Chr1 | 15227828 | 15227829 | 1 | |
| Deletion | Chr1 | 15807036 | 15807041 | 5 | |
| Deletion | Chr11 | 16606113 | 16606122 | 9 | |
| Deletion | Chr4 | 17197780 | 17197782 | 2 | |
| Deletion | Chr6 | 17913709 | 17913710 | 1 | |
| Deletion | Chr3 | 19009001 | 19009013 | 12 | |
| Deletion | Chr7 | 20518301 | 20518350 | 49 | |
| Deletion | Chr5 | 21963316 | 21963321 | 5 | |
| Deletion | Chr10 | 22174105 | 22174110 | 5 | |
| Deletion | Chr9 | 22892231 | 22892232 | 1 | |
| Deletion | Chr5 | 22922439 | 22923060 | 621 | |
| Deletion | Chr12 | 26252926 | 26252945 | 19 | |
| Deletion | Chr2 | 27241890 | 27241891 | 1 | LOC_Os02g44975 |
| Deletion | Chr5 | 27770559 | 27770566 | 7 | |
| Deletion | Chr3 | 28442153 | 28442154 | 1 | |
| Deletion | Chr7 | 29495747 | 29495748 | 1 | |
| Deletion | Chr1 | 37279013 | 37279029 | 16 | LOC_Os01g64190 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23960258 | 23960263 | 6 | LOC_Os01g42270 |
| Insertion | Chr1 | 29615782 | 29615797 | 16 | |
| Insertion | Chr1 | 5223729 | 5223729 | 1 | |
| Insertion | Chr10 | 6182237 | 6182237 | 1 | |
| Insertion | Chr10 | 7629684 | 7629687 | 4 | |
| Insertion | Chr12 | 24348928 | 24348928 | 1 | |
| Insertion | Chr2 | 1864451 | 1864451 | 1 | LOC_Os02g04240 |
| Insertion | Chr4 | 32903332 | 32903332 | 1 | |
| Insertion | Chr5 | 12017251 | 12017271 | 21 | |
| Insertion | Chr6 | 5201317 | 5201318 | 2 | |
| Insertion | Chr6 | 5767785 | 5767798 | 14 | |
| Insertion | Chr7 | 4124573 | 4124574 | 2 | |
| Insertion | Chr9 | 11057437 | 11057465 | 29 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 11834017 | 29552990 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 5281345 | Chr7 | 5459882 | |
| Translocation | Chr9 | 14643606 | Chr7 | 2367382 | |
| Translocation | Chr10 | 15057021 | Chr3 | 14226468 | |
| Translocation | Chr10 | 15057823 | Chr3 | 14316455 | LOC_Os03g25070 |
| Translocation | Chr4 | 23739020 | Chr3 | 6420341 | 2 |
| Translocation | Chr4 | 23739026 | Chr3 | 6412010 | LOC_Os04g39864 |
