Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1634-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1634-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 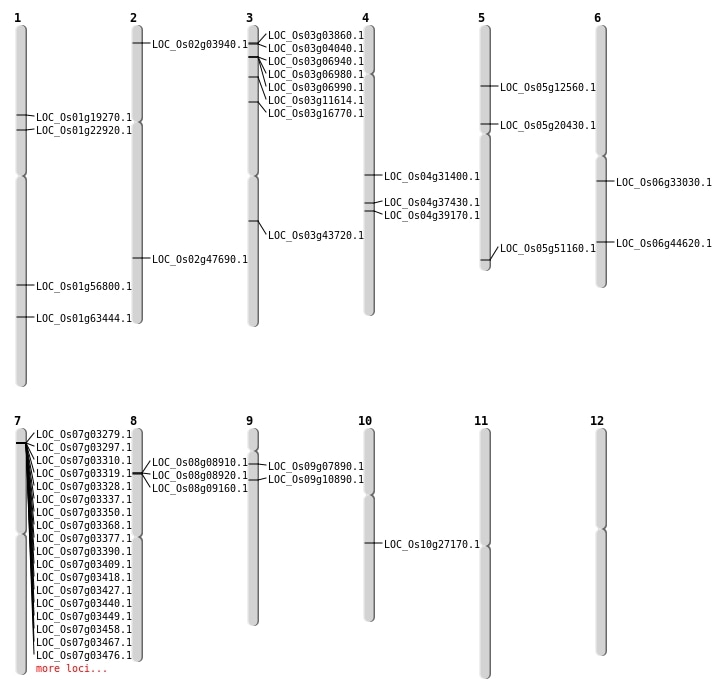
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12885196 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g22920 |
| SBS | Chr1 | 12885197 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g22920 |
| SBS | Chr1 | 32783665 | G-A | HET | START_GAINED | LOC_Os01g56800 |
| SBS | Chr1 | 34856725 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 36757344 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63444 |
| SBS | Chr10 | 15329998 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 15906346 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15906347 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16678289 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 649603 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 17756787 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27667797 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 29161015 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g47690 |
| SBS | Chr2 | 33789179 | T-A | HET | INTRON | |
| SBS | Chr2 | 4252267 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 17224898 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 17762343 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1844126 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04040 |
| SBS | Chr4 | 1836036 | T-A | HET | INTRON | |
| SBS | Chr4 | 5384199 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5920782 | G-A | HET | INTRON | |
| SBS | Chr4 | 6345537 | G-A | HET | INTRON | |
| SBS | Chr5 | 11988951 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 18978342 | T-A | Homo | INTERGENIC | |
| SBS | Chr5 | 19472537 | G-C | Homo | INTRON | |
| SBS | Chr5 | 23542626 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 26631103 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 7208984 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g12560 |
| SBS | Chr5 | 8950485 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 10472670 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 23037107 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2804514 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr6 | 28435598 | T-C | HET | INTRON | |
| SBS | Chr6 | 29709042 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 14340894 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 4771348 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 10751531 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 12285357 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 23108300 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 25696213 | T-A | HET | INTRON | |
| SBS | Chr9 | 2890704 | T-C | HET | INTRON | |
| SBS | Chr9 | 8107003 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8510921 | T-G | HET | UTR_3_PRIME |
Deletions: 45
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 53848 | 53850 | 2 | |
| Deletion | Chr8 | 536100 | 536101 | 1 | |
| Deletion | Chr12 | 1249658 | 1249661 | 3 | |
| Deletion | Chr7 | 1319001 | 1411000 | 92000 | 21 |
| Deletion | Chr3 | 1576025 | 1576038 | 13 | |
| Deletion | Chr3 | 1734737 | 1743739 | 9002 | LOC_Os03g03860 |
| Deletion | Chr11 | 2204136 | 2204150 | 14 | |
| Deletion | Chr7 | 3048001 | 3105000 | 57000 | 9 |
| Deletion | Chr3 | 3560627 | 3566984 | 6357 | 2 |
| Deletion | Chr9 | 3764679 | 3764691 | 12 | |
| Deletion | Chr9 | 4011896 | 4011898 | 2 | LOC_Os09g07890 |
| Deletion | Chr9 | 4772102 | 4772103 | 1 | |
| Deletion | Chr4 | 4774335 | 4774341 | 6 | |
| Deletion | Chr8 | 5180813 | 5189460 | 8647 | 2 |
| Deletion | Chr8 | 5312461 | 5312463 | 2 | LOC_Os08g09160 |
| Deletion | Chr9 | 5973566 | 5973567 | 1 | LOC_Os09g10890 |
| Deletion | Chr3 | 6050504 | 6057752 | 7248 | LOC_Os03g11614 |
| Deletion | Chr4 | 8439565 | 8439566 | 1 | |
| Deletion | Chr3 | 9295058 | 9295062 | 4 | LOC_Os03g16770 |
| Deletion | Chr2 | 9443849 | 9443850 | 1 | |
| Deletion | Chr1 | 11019110 | 11019118 | 8 | |
| Deletion | Chr2 | 12213236 | 12213237 | 1 | |
| Deletion | Chr8 | 12666498 | 12666513 | 15 | |
| Deletion | Chr12 | 13229311 | 13229327 | 16 | |
| Deletion | Chr4 | 13598829 | 13598831 | 2 | |
| Deletion | Chr4 | 14515072 | 14515073 | 1 | |
| Deletion | Chr10 | 14827428 | 14827439 | 11 | |
| Deletion | Chr8 | 15048170 | 15048171 | 1 | |
| Deletion | Chr10 | 15906249 | 15906255 | 6 | |
| Deletion | Chr11 | 16315983 | 16315985 | 2 | |
| Deletion | Chr10 | 16419872 | 16419884 | 12 | |
| Deletion | Chr6 | 17335844 | 17335850 | 6 | |
| Deletion | Chr6 | 17499979 | 17499982 | 3 | |
| Deletion | Chr6 | 19227474 | 19227479 | 5 | LOC_Os06g33030 |
| Deletion | Chr1 | 19739872 | 19739881 | 9 | |
| Deletion | Chr1 | 19851610 | 19851611 | 1 | |
| Deletion | Chr4 | 22080967 | 22080973 | 6 | |
| Deletion | Chr11 | 24348487 | 24348500 | 13 | |
| Deletion | Chr3 | 24449554 | 24449555 | 1 | LOC_Os03g43720 |
| Deletion | Chr11 | 25833282 | 25833284 | 2 | |
| Deletion | Chr3 | 26522806 | 26522808 | 2 | |
| Deletion | Chr2 | 27390355 | 27390356 | 1 | |
| Deletion | Chr3 | 28517266 | 28517296 | 30 | |
| Deletion | Chr4 | 35212755 | 35212766 | 11 | |
| Deletion | Chr1 | 42914797 | 42914800 | 3 |
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42913055 | 42913055 | 1 | |
| Insertion | Chr1 | 8434486 | 8434547 | 62 | |
| Insertion | Chr10 | 7497182 | 7497183 | 2 | |
| Insertion | Chr11 | 3168493 | 3168510 | 18 | |
| Insertion | Chr11 | 4878367 | 4878368 | 2 | |
| Insertion | Chr12 | 9729905 | 9729906 | 2 | |
| Insertion | Chr2 | 23649329 | 23649356 | 28 | |
| Insertion | Chr3 | 16289691 | 16289692 | 2 | |
| Insertion | Chr3 | 16854817 | 16854818 | 2 | |
| Insertion | Chr3 | 18369894 | 18369970 | 77 | |
| Insertion | Chr3 | 25392671 | 25392671 | 1 | |
| Insertion | Chr4 | 18779650 | 18779658 | 9 | LOC_Os04g31400 |
| Insertion | Chr4 | 26610764 | 26610764 | 1 | |
| Insertion | Chr4 | 30028690 | 30028693 | 4 | |
| Insertion | Chr4 | 9469462 | 9469462 | 1 | |
| Insertion | Chr5 | 16603496 | 16603513 | 18 | |
| Insertion | Chr5 | 29348402 | 29348402 | 1 | LOC_Os05g51160 |
| Insertion | Chr6 | 19505216 | 19505216 | 1 | |
| Insertion | Chr6 | 26612108 | 26612131 | 24 | |
| Insertion | Chr6 | 26943244 | 26943244 | 1 | LOC_Os06g44620 |
| Insertion | Chr7 | 8457698 | 8457709 | 12 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 14322278 | 14626660 | LOC_Os10g27170 |
| Inversion | Chr10 | 14322317 | 14626669 | LOC_Os10g27170 |
| Inversion | Chr11 | 17399199 | 17877199 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 3524178 | Chr1 | 629528 | |
| Translocation | Chr3 | 3527694 | Chr1 | 629486 | LOC_Os03g06940 |
| Translocation | Chr8 | 5180827 | Chr7 | 14210904 | LOC_Os08g08910 |
| Translocation | Chr8 | 5189457 | Chr1 | 10897036 | LOC_Os01g19270 |
| Translocation | Chr8 | 5189504 | Chr7 | 14210903 | |
| Translocation | Chr3 | 6050513 | Chr2 | 1675802 | LOC_Os02g03940 |
| Translocation | Chr3 | 6057732 | Chr2 | 1675808 | 2 |
| Translocation | Chr7 | 14210902 | Chr1 | 10896698 | LOC_Os01g19270 |
| Translocation | Chr4 | 22308947 | Chr3 | 1734738 | LOC_Os04g37430 |
| Translocation | Chr4 | 23295106 | Chr3 | 1743776 | LOC_Os04g39170 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 1714089 | 3566935 | 1852846 |
