Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1650-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1650-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 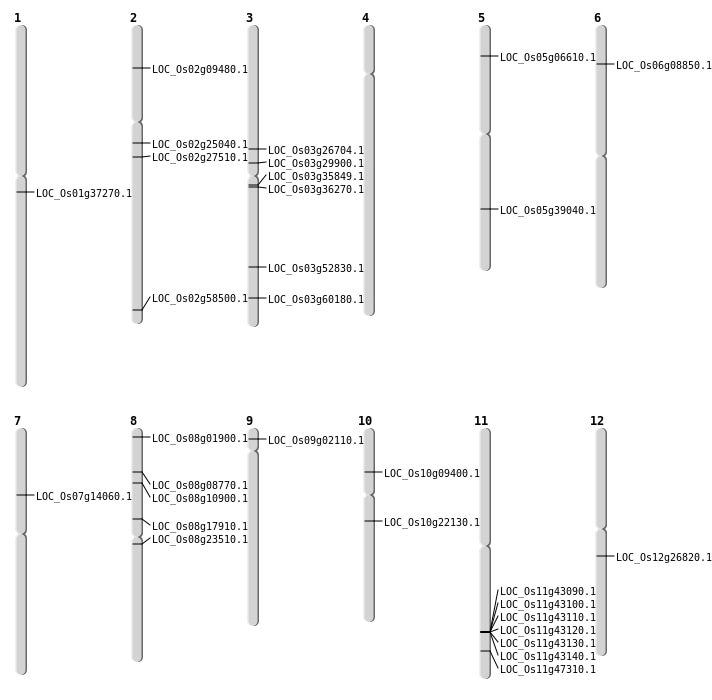
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10092188 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 18736798 | C-A | Homo | INTRON | |
| SBS | Chr1 | 20143596 | T-C | Homo | INTERGENIC | |
| SBS | Chr1 | 26252860 | T-C | HET | INTRON | |
| SBS | Chr1 | 28470221 | C-T | HET | INTRON | |
| SBS | Chr1 | 29195750 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 3512294 | A-G | HET | INTRON | |
| SBS | Chr10 | 11429651 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g22130 |
| SBS | Chr10 | 5074124 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g09400 |
| SBS | Chr10 | 8411526 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 4131781 | C-T | Homo | INTERGENIC | |
| SBS | Chr11 | 6919815 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 7588458 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 15708415 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 27684006 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 28612817 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 30949176 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35771204 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g58500 |
| SBS | Chr2 | 35771205 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 12419285 | G-T | Homo | INTERGENIC | |
| SBS | Chr3 | 13559330 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 17032675 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g29900 |
| SBS | Chr3 | 19866781 | A-T | HET | START_GAINED | LOC_Os03g35849 |
| SBS | Chr3 | 23492601 | C-G | Homo | UTR_5_PRIME | |
| SBS | Chr3 | 27907261 | G-A | HET | INTRON | |
| SBS | Chr3 | 34227901 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g60180 |
| SBS | Chr3 | 36371977 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23765167 | G-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr4 | 30148952 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 11210714 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 16735054 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 1828370 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 22900574 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g39040 |
| SBS | Chr5 | 29687009 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8121865 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 8121867 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2546873 | C-A | Homo | UTR_5_PRIME | |
| SBS | Chr6 | 4437310 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g08850 |
| SBS | Chr6 | 9910664 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 15155308 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 20923497 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 3988063 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 8020454 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g14060 |
| SBS | Chr8 | 13524985 | T-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 20001320 | C-T | HET | INTRON | |
| SBS | Chr8 | 2621877 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 10524645 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13151096 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 4334476 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 7843117 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 790815 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g02110 |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 555817 | 555829 | 12 | LOC_Os08g01900 |
| Deletion | Chr11 | 2138983 | 2138990 | 7 | |
| Deletion | Chr11 | 2809592 | 2809623 | 31 | |
| Deletion | Chr5 | 3412152 | 3412157 | 5 | LOC_Os05g06610 |
| Deletion | Chr2 | 4869246 | 4869247 | 1 | LOC_Os02g09480 |
| Deletion | Chr6 | 4920979 | 4920982 | 3 | |
| Deletion | Chr8 | 5085662 | 5085665 | 3 | LOC_Os08g08770 |
| Deletion | Chr8 | 6428079 | 6428084 | 5 | LOC_Os08g10900 |
| Deletion | Chr10 | 7349824 | 7349834 | 10 | |
| Deletion | Chr5 | 10314538 | 10314539 | 1 | |
| Deletion | Chr8 | 11382016 | 11382017 | 1 | |
| Deletion | Chr11 | 12088883 | 12115598 | 26715 | |
| Deletion | Chr6 | 12654310 | 12654311 | 1 | |
| Deletion | Chr8 | 14237743 | 14237746 | 3 | LOC_Os08g23510 |
| Deletion | Chr2 | 14519816 | 14519824 | 8 | LOC_Os02g25040 |
| Deletion | Chr4 | 15111427 | 15111428 | 1 | |
| Deletion | Chr4 | 15582141 | 15582151 | 10 | |
| Deletion | Chr12 | 15711844 | 15711846 | 2 | LOC_Os12g26820 |
| Deletion | Chr4 | 15744792 | 15744802 | 10 | |
| Deletion | Chr2 | 16283558 | 16283569 | 11 | LOC_Os02g27510 |
| Deletion | Chr6 | 17319242 | 17319244 | 2 | |
| Deletion | Chr2 | 20510035 | 20510038 | 3 | |
| Deletion | Chr1 | 20813836 | 20813838 | 2 | LOC_Os01g37270 |
| Deletion | Chr8 | 20947546 | 20947557 | 11 | |
| Deletion | Chr10 | 21747312 | 21747313 | 1 | |
| Deletion | Chr4 | 24198643 | 24198652 | 9 | |
| Deletion | Chr12 | 24522921 | 24522926 | 5 | |
| Deletion | Chr6 | 25556332 | 25556334 | 2 | |
| Deletion | Chr11 | 25992001 | 26028000 | 36000 | 6 |
| Deletion | Chr2 | 29515176 | 29515181 | 5 | |
| Deletion | Chr3 | 30293103 | 30293106 | 3 | LOC_Os03g52830 |
| Deletion | Chr2 | 33671974 | 33672020 | 46 | |
| Deletion | Chr3 | 35653844 | 35653847 | 3 | |
| Deletion | Chr1 | 40578244 | 40578247 | 3 | |
| Deletion | Chr1 | 42866990 | 42866993 | 3 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 18534196 | 18534197 | 2 | |
| Insertion | Chr2 | 33018962 | 33018963 | 2 | |
| Insertion | Chr3 | 20120884 | 20120885 | 2 | LOC_Os03g36270 |
| Insertion | Chr3 | 25693159 | 25693160 | 2 | |
| Insertion | Chr3 | 365270 | 365270 | 1 | |
| Insertion | Chr4 | 31713913 | 31713915 | 3 | |
| Insertion | Chr7 | 22131048 | 22131052 | 5 | |
| Insertion | Chr8 | 21320636 | 21320685 | 50 | |
| Insertion | Chr8 | 8336123 | 8336124 | 2 | |
| Insertion | Chr9 | 19514508 | 19514508 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 1919887 | 19701008 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 4961285 | Chr7 | 3359558 | |
| Translocation | Chr11 | 5070069 | Chr10 | 7556024 | |
| Translocation | Chr8 | 10991578 | Chr3 | 15245749 | 2 |
| Translocation | Chr11 | 13614688 | Chr4 | 15843 | |
| Translocation | Chr6 | 13827061 | Chr3 | 31363407 | |
| Translocation | Chr11 | 28452495 | Chr4 | 22510681 | LOC_Os11g47310 |
