Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1660-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1660-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 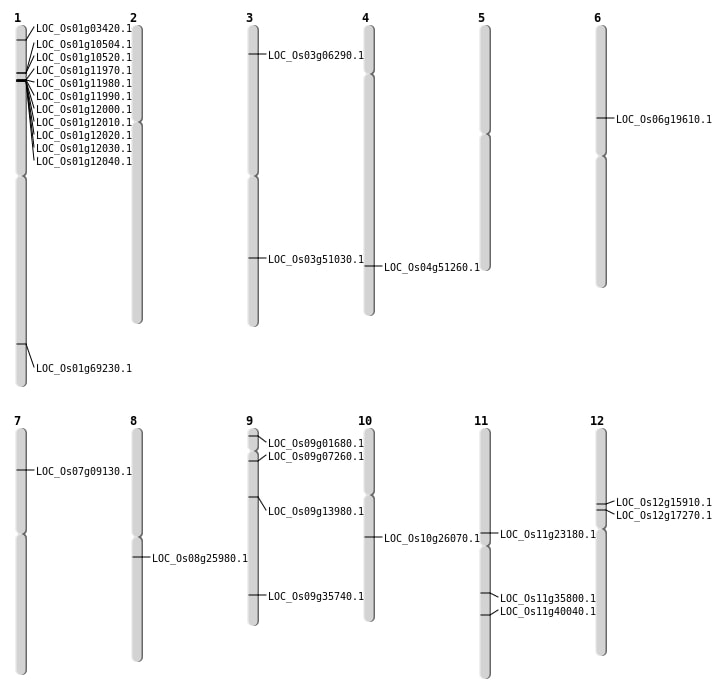
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19062047 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 40236435 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g69230 |
| SBS | Chr10 | 13502029 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26070 |
| SBS | Chr11 | 11493372 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 13346466 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23180 |
| SBS | Chr11 | 13696896 | A-T | HET | INTRON | |
| SBS | Chr11 | 21011729 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35800 |
| SBS | Chr11 | 5645707 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr12 | 10762243 | C-A | HET | INTRON | |
| SBS | Chr12 | 14304436 | A-T | Homo | INTERGENIC | |
| SBS | Chr12 | 23163029 | A-G | HET | INTRON | |
| SBS | Chr12 | 9056295 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15910 |
| SBS | Chr2 | 14744257 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15995611 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 11914889 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 22806477 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 29171496 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51030 |
| SBS | Chr3 | 3151860 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06290 |
| SBS | Chr3 | 9416377 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 13136832 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 16618361 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16618362 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21553656 | A-T | HET | INTRON | |
| SBS | Chr4 | 23800004 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23908116 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 25834734 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 28824811 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 4664314 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 2987508 | C-A | HET | INTRON | |
| SBS | Chr6 | 1216143 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15154401 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 21778151 | A-G | HET | INTRON | |
| SBS | Chr6 | 26501757 | A-G | Homo | INTERGENIC | |
| SBS | Chr6 | 5434705 | G-T | Homo | INTERGENIC | |
| SBS | Chr6 | 5888046 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21461588 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 26938426 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 15808409 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g25980 |
| SBS | Chr8 | 24087856 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 24087857 | C-A | Homo | INTERGENIC | |
| SBS | Chr8 | 5889593 | T-C | HET | INTRON | |
| SBS | Chr9 | 17975533 | T-G | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 352743 | 352747 | 4 | |
| Deletion | Chr8 | 465264 | 465404 | 140 | |
| Deletion | Chr11 | 934743 | 934744 | 1 | |
| Deletion | Chr1 | 1375553 | 1375564 | 11 | LOC_Os01g03420 |
| Deletion | Chr9 | 3560610 | 3560633 | 23 | LOC_Os09g07260 |
| Deletion | Chr1 | 5554798 | 5583469 | 28671 | 2 |
| Deletion | Chr2 | 5805641 | 5805642 | 1 | |
| Deletion | Chr1 | 6521132 | 6549964 | 28832 | 8 |
| Deletion | Chr9 | 8234866 | 8235001 | 135 | LOC_Os09g13980 |
| Deletion | Chr2 | 8693974 | 8693976 | 2 | |
| Deletion | Chr7 | 8962845 | 8962847 | 2 | |
| Deletion | Chr6 | 9495787 | 9495788 | 1 | |
| Deletion | Chr12 | 9883714 | 9883733 | 19 | LOC_Os12g17270 |
| Deletion | Chr11 | 10193082 | 10193083 | 1 | |
| Deletion | Chr2 | 13112086 | 13112088 | 2 | |
| Deletion | Chr12 | 14565436 | 14565437 | 1 | |
| Deletion | Chr4 | 15467932 | 15468095 | 163 | |
| Deletion | Chr1 | 15689146 | 15689147 | 1 | |
| Deletion | Chr5 | 15904703 | 15904704 | 1 | |
| Deletion | Chr9 | 16951638 | 16951639 | 1 | |
| Deletion | Chr9 | 17151533 | 17151534 | 1 | |
| Deletion | Chr12 | 17501400 | 17501411 | 11 | |
| Deletion | Chr12 | 19633468 | 19633469 | 1 | |
| Deletion | Chr8 | 20202193 | 20202207 | 14 | |
| Deletion | Chr9 | 20558206 | 20558207 | 1 | LOC_Os09g35740 |
| Deletion | Chr11 | 23874735 | 23874739 | 4 | LOC_Os11g40040 |
| Deletion | Chr12 | 24302505 | 24302507 | 2 | |
| Deletion | Chr12 | 24361140 | 24361153 | 13 | |
| Deletion | Chr2 | 25200159 | 25200160 | 1 | |
| Deletion | Chr2 | 26043189 | 26043190 | 1 | |
| Deletion | Chr7 | 26072016 | 26072017 | 1 | |
| Deletion | Chr1 | 26262240 | 26262248 | 8 | |
| Deletion | Chr11 | 28439782 | 28439943 | 161 | |
| Deletion | Chr2 | 35847890 | 35847892 | 2 |
Insertions: 8
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 18655811 | 19268779 | |
| Inversion | Chr4 | 30373114 | 30616851 | LOC_Os04g51260 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 488389 | Chr6 | 11179783 | 2 |
| Translocation | Chr9 | 488420 | Chr6 | 11179452 | LOC_Os09g01680 |
| Translocation | Chr12 | 27018183 | Chr7 | 4769691 | LOC_Os07g09130 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr1 | 6535298 | 6535298 | 0 | LOC_Os01g12000 |
