Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1669-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1669-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 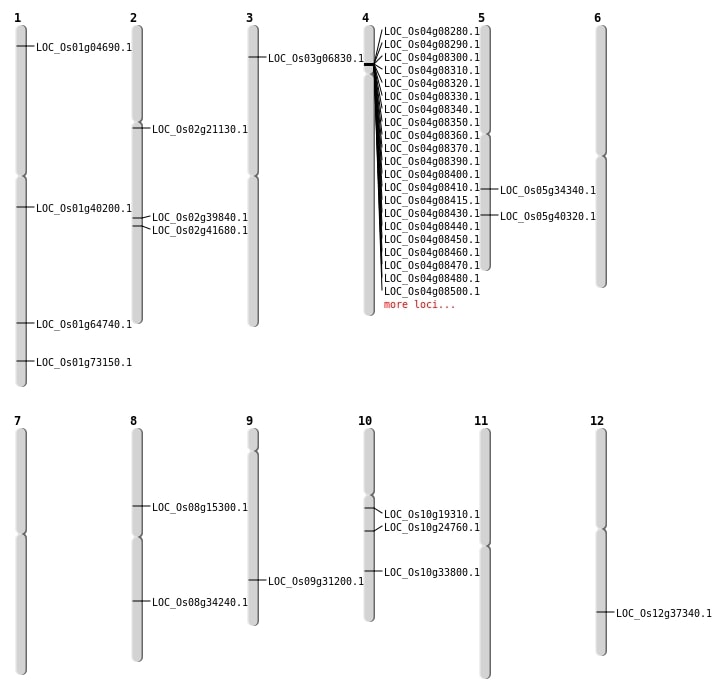
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11086863 | C-A | HET | ||
| SBS | Chr1 | 27979975 | G-T | Homo | ||
| SBS | Chr1 | 8088859 | A-T | HET | ||
| SBS | Chr10 | 10600041 | G-T | Homo | ||
| SBS | Chr10 | 15398045 | G-A | Homo | ||
| SBS | Chr11 | 11037880 | C-T | Homo | ||
| SBS | Chr11 | 13740429 | C-T | HET | ||
| SBS | Chr11 | 23229216 | A-C | HET | ||
| SBS | Chr12 | 22929920 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37340 |
| SBS | Chr12 | 23443526 | G-T | HET | ||
| SBS | Chr12 | 24450866 | G-C | Homo | ||
| SBS | Chr2 | 15428227 | C-T | HET | ||
| SBS | Chr2 | 2677785 | C-T | Homo | ||
| SBS | Chr3 | 19367332 | G-T | Homo | ||
| SBS | Chr3 | 35072705 | G-T | HET | ||
| SBS | Chr3 | 7557776 | A-G | Homo | ||
| SBS | Chr3 | 9926848 | G-A | Homo | ||
| SBS | Chr4 | 32899185 | G-T | HET | ||
| SBS | Chr4 | 33608480 | G-T | HET | ||
| SBS | Chr4 | 7150768 | A-T | HET | ||
| SBS | Chr4 | 7346744 | C-T | HET | ||
| SBS | Chr5 | 27261187 | G-T | HET | ||
| SBS | Chr7 | 25010996 | G-A | HET | ||
| SBS | Chr8 | 23088073 | T-A | Homo | ||
| SBS | Chr8 | 7270493 | A-T | Homo |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 3552401 | 3552402 | 1 | |
| Deletion | Chr4 | 4421001 | 4619000 | 197999 | 23 |
| Deletion | Chr9 | 4486862 | 4486868 | 6 | |
| Deletion | Chr4 | 4629001 | 4748000 | 118999 | 20 |
| Deletion | Chr1 | 9032805 | 9032806 | 1 | |
| Deletion | Chr10 | 15568932 | 15568948 | 16 | |
| Deletion | Chr8 | 15839272 | 15839285 | 13 | |
| Deletion | Chr1 | 17232530 | 17232531 | 1 | |
| Deletion | Chr9 | 17298477 | 17298478 | 1 | |
| Deletion | Chr5 | 20355402 | 20355404 | 2 | LOC_Os05g34340 |
| Deletion | Chr1 | 21207069 | 21207070 | 1 | |
| Deletion | Chr8 | 21467812 | 21467814 | 2 | LOC_Os08g34240 |
| Deletion | Chr2 | 23462995 | 23462996 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 14418717 | 14418717 | 1 | |
| Insertion | Chr7 | 2860482 | 2860483 | 2 |
Inversions: 24
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 2108818 | 2109151 | 2 |
| Inversion | Chr3 | 3278795 | 3279141 | |
| Inversion | Chr3 | 3462921 | 3463131 | LOC_Os03g06830 |
| Inversion | Chr1 | 3642688 | 3642943 | |
| Inversion | Chr7 | 4328513 | 4328727 | |
| Inversion | Chr10 | 9840597 | 9841208 | LOC_Os10g19310 |
| Inversion | Chr4 | 11026197 | 11026322 | |
| Inversion | Chr9 | 12234791 | 12235003 | |
| Inversion | Chr2 | 12545708 | 12545978 | LOC_Os02g21130 |
| Inversion | Chr3 | 12912746 | 12912881 | |
| Inversion | Chr11 | 14269426 | 14269642 | |
| Inversion | Chr10 | 17916850 | 17917250 | LOC_Os10g33800 |
| Inversion | Chr7 | 18305797 | 18305978 | |
| Inversion | Chr9 | 18761345 | 18761818 | LOC_Os09g31200 |
| Inversion | Chr10 | 19042382 | 19042566 | |
| Inversion | Chr10 | 21681890 | 21682213 | |
| Inversion | Chr10 | 22195985 | 22196273 | |
| Inversion | Chr1 | 22683747 | 22683911 | LOC_Os01g40200 |
| Inversion | Chr5 | 23693813 | 23693997 | LOC_Os05g40320 |
| Inversion | Chr2 | 24073780 | 24074064 | LOC_Os02g39840 |
| Inversion | Chr2 | 25008262 | 25008589 | LOC_Os02g41680 |
| Inversion | Chr4 | 32703254 | 32703780 | 2 |
| Inversion | Chr1 | 37563750 | 37564037 | LOC_Os01g64740 |
| Inversion | Chr1 | 42422808 | 42423174 | LOC_Os01g73150 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 12733718 | Chr8 | 9303483 | 2 |
| Translocation | Chr4 | 32040721 | Chr3 | 31809571 |
