Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1679-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1679-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 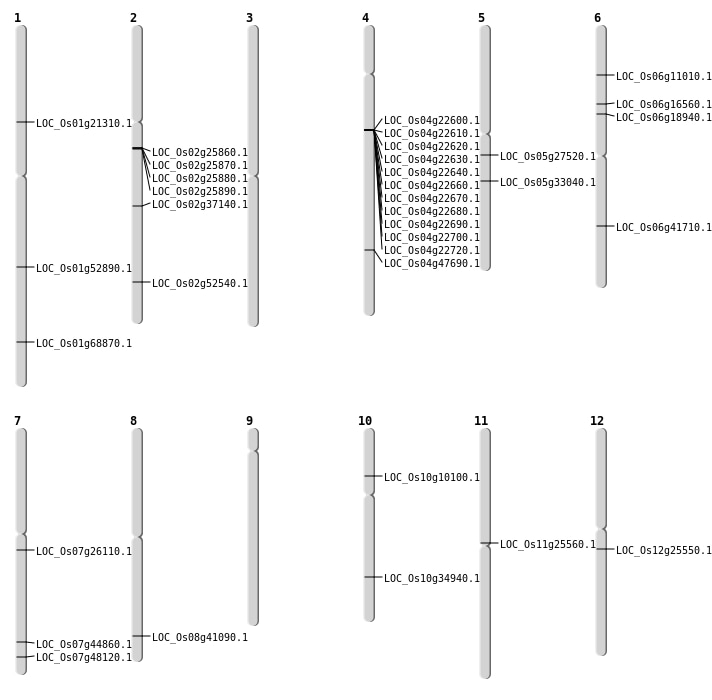
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16181107 | A-G | HET | ||
| SBS | Chr1 | 17054878 | A-G | Homo | ||
| SBS | Chr1 | 174845 | G-A | HET | ||
| SBS | Chr10 | 3758576 | A-T | Homo | ||
| SBS | Chr10 | 6406642 | G-C | Homo | ||
| SBS | Chr11 | 16012961 | A-C | Homo | ||
| SBS | Chr11 | 23101178 | G-A | Homo | ||
| SBS | Chr2 | 26419856 | A-G | HET | ||
| SBS | Chr3 | 27909180 | C-T | HET | ||
| SBS | Chr4 | 23304507 | C-A | Homo | ||
| SBS | Chr4 | 5822614 | T-C | Homo | ||
| SBS | Chr5 | 10729452 | G-A | Homo | ||
| SBS | Chr5 | 12508210 | C-A | HET | ||
| SBS | Chr5 | 16008844 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g27520 |
| SBS | Chr5 | 23493360 | G-T | Homo | ||
| SBS | Chr5 | 28785601 | A-T | Homo | ||
| SBS | Chr5 | 7397978 | T-C | Homo | ||
| SBS | Chr6 | 21700344 | T-C | Homo | ||
| SBS | Chr7 | 14347612 | G-C | Homo | ||
| SBS | Chr7 | 22804080 | T-A | HET | ||
| SBS | Chr7 | 26539910 | G-A | HET | ||
| SBS | Chr7 | 26539916 | C-A | HET | ||
| SBS | Chr7 | 27673686 | G-A | HET | ||
| SBS | Chr7 | 8585143 | A-C | Homo | ||
| SBS | Chr8 | 10866412 | T-C | HET | ||
| SBS | Chr8 | 18526655 | C-T | Homo | ||
| SBS | Chr8 | 19947571 | G-A | HET | ||
| SBS | Chr9 | 12522160 | G-A | HET | ||
| SBS | Chr9 | 17866125 | A-C | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1660892 | 1660900 | 8 | |
| Deletion | Chr9 | 3835582 | 3835585 | 3 | |
| Deletion | Chr1 | 8276449 | 8276457 | 8 | |
| Deletion | Chr6 | 12447510 | 12447520 | 10 | |
| Deletion | Chr4 | 12800001 | 12886000 | 85999 | 11 |
| Deletion | Chr3 | 14574457 | 14574477 | 20 | |
| Deletion | Chr12 | 14785741 | 14785742 | 1 | LOC_Os12g25550 |
| Deletion | Chr2 | 15153001 | 15189000 | 35999 | 4 |
| Deletion | Chr11 | 16360904 | 16360914 | 10 | |
| Deletion | Chr3 | 18497510 | 18497517 | 7 | |
| Deletion | Chr10 | 19043169 | 19043175 | 6 | |
| Deletion | Chr9 | 19479611 | 19479612 | 1 | |
| Deletion | Chr6 | 19630922 | 19630928 | 6 | |
| Deletion | Chr2 | 21009364 | 21009370 | 6 | |
| Deletion | Chr11 | 22479923 | 22479928 | 5 | |
| Deletion | Chr7 | 23377643 | 23377645 | 2 | |
| Deletion | Chr3 | 23762801 | 23762824 | 23 | |
| Deletion | Chr2 | 23925598 | 23925601 | 3 | |
| Deletion | Chr8 | 25972874 | 25972890 | 16 | LOC_Os08g41090 |
| Deletion | Chr7 | 26655546 | 26655562 | 16 | |
| Deletion | Chr1 | 40022824 | 40022828 | 4 | LOC_Os01g68870 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 31582346 | 31582347 | 2 |
Inversions: 31
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 590292 | 590599 | |
| Inversion | Chr12 | 3828054 | 3828247 | |
| Inversion | Chr6 | 4866414 | 4866697 | |
| Inversion | Chr1 | 5362871 | 5363107 | |
| Inversion | Chr10 | 5516391 | 5516638 | LOC_Os10g10100 |
| Inversion | Chr6 | 5760745 | 5761118 | LOC_Os06g11010 |
| Inversion | Chr3 | 8076092 | 8076696 | |
| Inversion | Chr6 | 9496312 | 9496555 | LOC_Os06g16560 |
| Inversion | Chr7 | 10209382 | 10209577 | |
| Inversion | Chr6 | 10766144 | 10766306 | LOC_Os06g18940 |
| Inversion | Chr1 | 11906510 | 11906765 | LOC_Os01g21310 |
| Inversion | Chr3 | 13190752 | 13191168 | |
| Inversion | Chr11 | 14569106 | 14569420 | LOC_Os11g25560 |
| Inversion | Chr7 | 14989225 | 14989488 | LOC_Os07g26110 |
| Inversion | Chr10 | 18657719 | 18657985 | LOC_Os10g34940 |
| Inversion | Chr5 | 19366859 | 19366989 | LOC_Os05g33040 |
| Inversion | Chr5 | 20767939 | 20768047 | |
| Inversion | Chr12 | 22071692 | 22071881 | |
| Inversion | Chr2 | 22452845 | 22453058 | LOC_Os02g37140 |
| Inversion | Chr1 | 24703559 | 24703736 | |
| Inversion | Chr6 | 24996954 | 24997321 | LOC_Os06g41710 |
| Inversion | Chr3 | 25245278 | 25245570 | |
| Inversion | Chr7 | 26784244 | 26784448 | LOC_Os07g44860 |
| Inversion | Chr2 | 27396098 | 27396462 | |
| Inversion | Chr8 | 27648249 | 27648639 | |
| Inversion | Chr4 | 28294103 | 28294393 | LOC_Os04g47690 |
| Inversion | Chr7 | 28738941 | 28739198 | LOC_Os07g48120 |
| Inversion | Chr2 | 30320568 | 30320833 | |
| Inversion | Chr2 | 32150172 | 32150400 | LOC_Os02g52540 |
| Inversion | Chr4 | 32853480 | 32853845 | |
| Inversion | Chr1 | 34189590 | 34189771 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 12800477 | Chr1 | 30398590 | LOC_Os04g22600 |
| Translocation | Chr4 | 12801588 | Chr1 | 30413013 | 2 |
| Translocation | Chr8 | 13370121 | Chr4 | 2808015 | |
| Translocation | Chr7 | 17709928 | Chr1 | 30990138 |
