Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1681-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1681-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 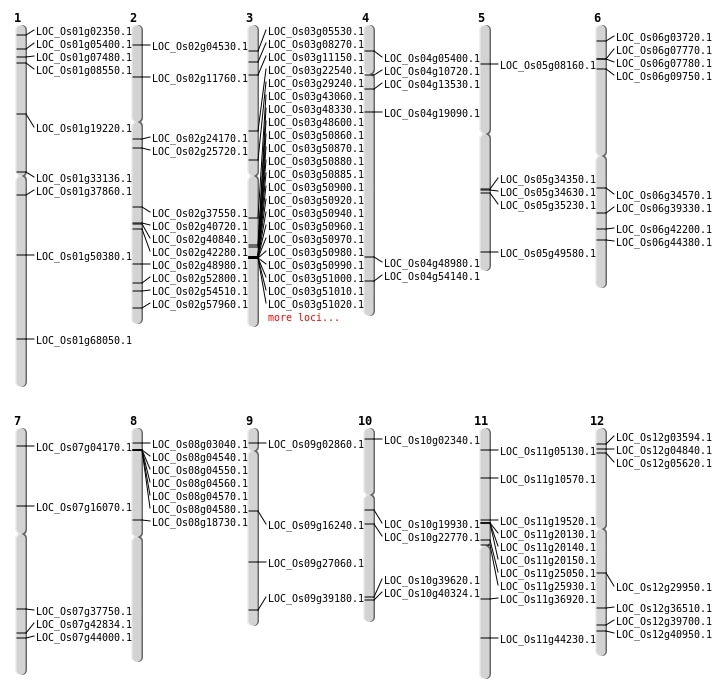
|
| Variant Information |
|---|
Single base substitutions: 13
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30731820 | C-T | Homo | ||
| SBS | Chr10 | 22014883 | C-A | Homo | ||
| SBS | Chr3 | 20440096 | C-T | Homo | ||
| SBS | Chr3 | 21813942 | C-T | Homo | ||
| SBS | Chr3 | 7796573 | T-A | Homo | ||
| SBS | Chr4 | 13564858 | G-A | Homo | ||
| SBS | Chr4 | 19777305 | T-C | Homo | ||
| SBS | Chr5 | 818597 | A-T | Homo | ||
| SBS | Chr5 | 8468542 | C-G | Homo | ||
| SBS | Chr6 | 13992667 | C-A | HET | ||
| SBS | Chr6 | 1721917 | T-C | Homo | ||
| SBS | Chr6 | 25334967 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g42200 |
| SBS | Chr9 | 16905094 | A-T | Homo |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2243001 | 2286000 | 42999 | 5 |
| Deletion | Chr6 | 6964017 | 6964021 | 4 | |
| Deletion | Chr5 | 9025758 | 9025760 | 2 | |
| Deletion | Chr8 | 11181878 | 11181879 | 1 | LOC_Os08g18730 |
| Deletion | Chr11 | 11625001 | 11646000 | 20999 | 3 |
| Deletion | Chr10 | 12601676 | 12601677 | 1 | |
| Deletion | Chr7 | 14559916 | 14559920 | 4 | |
| Deletion | Chr8 | 14721641 | 14721646 | 5 | |
| Deletion | Chr8 | 21430106 | 21430130 | 24 | |
| Deletion | Chr11 | 21780483 | 21780495 | 12 | LOC_Os11g36920 |
| Deletion | Chr10 | 22974931 | 22974946 | 15 | |
| Deletion | Chr3 | 29057001 | 29206000 | 148999 | 19 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 216802 | 216803 | 2 | |
| Insertion | Chr10 | 18026780 | 18026783 | 4 | |
| Insertion | Chr6 | 24860355 | 24860355 | 1 | |
| Insertion | Chr8 | 17058883 | 17058883 | 1 |
Inversions: 135
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 743182 | 743322 | LOC_Os01g02350 |
| Inversion | Chr10 | 838121 | 838451 | LOC_Os10g02340 |
| Inversion | Chr3 | 1161440 | 1161857 | |
| Inversion | Chr9 | 1341885 | 1342081 | LOC_Os09g02860 |
| Inversion | Chr8 | 1365042 | 1365369 | LOC_Os08g03040 |
| Inversion | Chr12 | 1437552 | 1437816 | LOC_Os12g03594 |
| Inversion | Chr6 | 1471861 | 1472190 | LOC_Os06g03720 |
| Inversion | Chr7 | 1784322 | 1784575 | LOC_Os07g04170 |
| Inversion | Chr2 | 1798919 | 1799210 | |
| Inversion | Chr12 | 1863821 | 1864091 | |
| Inversion | Chr2 | 2014093 | 2014340 | LOC_Os02g04530 |
| Inversion | Chr12 | 2084375 | 2084549 | LOC_Os12g04840 |
| Inversion | Chr11 | 2254772 | 2254943 | LOC_Os11g05130 |
| Inversion | Chr4 | 2386253 | 2386405 | |
| Inversion | Chr12 | 2585937 | 2586248 | LOC_Os12g05620 |
| Inversion | Chr4 | 2706290 | 2706473 | LOC_Os04g05400 |
| Inversion | Chr3 | 2748229 | 2748444 | LOC_Os03g05530 |
| Inversion | Chr1 | 3181121 | 3181444 | |
| Inversion | Chr8 | 3413595 | 3413781 | |
| Inversion | Chr1 | 3551029 | 3551238 | 2 |
| Inversion | Chr5 | 3778361 | 3778757 | |
| Inversion | Chr1 | 4160558 | 4160909 | |
| Inversion | Chr3 | 4209546 | 4210027 | LOC_Os03g08270 |
| Inversion | Chr1 | 4245361 | 4245820 | 2 |
| Inversion | Chr5 | 4448639 | 4448801 | LOC_Os05g08160 |
| Inversion | Chr6 | 4968669 | 4968924 | LOC_Os06g09750 |
| Inversion | Chr8 | 5065313 | 5065561 | |
| Inversion | Chr3 | 5260054 | 5260178 | |
| Inversion | Chr3 | 5542003 | 5542105 | |
| Inversion | Chr3 | 5747480 | 5748054 | LOC_Os03g11150 |
| Inversion | Chr11 | 5797435 | 5797628 | LOC_Os11g10570 |
| Inversion | Chr4 | 5826656 | 5827073 | LOC_Os04g10720 |
| Inversion | Chr2 | 6073594 | 6073940 | LOC_Os02g11760 |
| Inversion | Chr7 | 6376004 | 6376231 | |
| Inversion | Chr10 | 6491200 | 6491489 | |
| Inversion | Chr3 | 6712509 | 6712618 | |
| Inversion | Chr12 | 7501383 | 7501772 | |
| Inversion | Chr3 | 7716346 | 7716476 | |
| Inversion | Chr9 | 9337206 | 9337369 | |
| Inversion | Chr7 | 9369276 | 9369483 | LOC_Os07g16070 |
| Inversion | Chr8 | 9501784 | 9502040 | |
| Inversion | Chr9 | 9914983 | 9915136 | LOC_Os09g16240 |
| Inversion | Chr10 | 9994992 | 9995181 | LOC_Os10g19930 |
| Inversion | Chr4 | 10597445 | 10965151 | LOC_Os04g19090 |
| Inversion | Chr2 | 10813739 | 10814144 | |
| Inversion | Chr1 | 10872701 | 10872844 | LOC_Os01g19220 |
| Inversion | Chr11 | 11138847 | 11138964 | |
| Inversion | Chr11 | 11252026 | 11252323 | LOC_Os11g19520 |
| Inversion | Chr10 | 11836193 | 11836418 | LOC_Os10g22770 |
| Inversion | Chr5 | 12088756 | 12088950 | |
| Inversion | Chr3 | 12271034 | 12271176 | |
| Inversion | Chr6 | 12695561 | 12695892 | |
| Inversion | Chr3 | 12945703 | 12945812 | LOC_Os03g22540 |
| Inversion | Chr2 | 14001587 | 14001759 | LOC_Os02g24170 |
| Inversion | Chr11 | 14274426 | 14274570 | LOC_Os11g25050 |
| Inversion | Chr7 | 14614396 | 14614802 | |
| Inversion | Chr10 | 14780579 | 14781014 | |
| Inversion | Chr11 | 14780780 | 14780978 | |
| Inversion | Chr11 | 14793715 | 14793977 | LOC_Os11g25930 |
| Inversion | Chr2 | 15059225 | 15059343 | LOC_Os02g25720 |
| Inversion | Chr8 | 15389492 | 15389815 | |
| Inversion | Chr2 | 15737735 | 15738143 | |
| Inversion | Chr1 | 16329171 | 16329358 | |
| Inversion | Chr9 | 16462000 | 16462239 | LOC_Os09g27060 |
| Inversion | Chr3 | 16628827 | 16629178 | LOC_Os03g29240 |
| Inversion | Chr12 | 16971351 | 16971487 | |
| Inversion | Chr9 | 17037305 | 17037595 | |
| Inversion | Chr12 | 17902543 | 17903047 | LOC_Os12g29950 |
| Inversion | Chr5 | 18072585 | 18072788 | |
| Inversion | Chr1 | 18243513 | 18243980 | LOC_Os01g33136 |
| Inversion | Chr7 | 19124745 | 19124992 | |
| Inversion | Chr12 | 19723045 | 19723191 | |
| Inversion | Chr3 | 20047930 | 20048266 | |
| Inversion | Chr6 | 20108531 | 20108788 | LOC_Os06g34570 |
| Inversion | Chr5 | 20360108 | 20539104 | 2 |
| Inversion | Chr5 | 20360116 | 20539104 | 2 |
| Inversion | Chr5 | 20932893 | 20933002 | LOC_Os05g35230 |
| Inversion | Chr6 | 21044107 | 21044426 | |
| Inversion | Chr10 | 21173957 | 21174165 | LOC_Os10g39620 |
| Inversion | Chr1 | 21189559 | 21189946 | LOC_Os01g37860 |
| Inversion | Chr4 | 21253403 | 21253717 | |
| Inversion | Chr10 | 21602933 | 21603110 | LOC_Os10g40324 |
| Inversion | Chr12 | 22350509 | 22350869 | LOC_Os12g36510 |
| Inversion | Chr9 | 22505510 | 22505637 | LOC_Os09g39180 |
| Inversion | Chr2 | 22558384 | 22558705 | |
| Inversion | Chr2 | 22655481 | 22655639 | 2 |
| Inversion | Chr6 | 23334648 | 23334828 | LOC_Os06g39330 |
| Inversion | Chr5 | 23764980 | 23765126 | |
| Inversion | Chr3 | 24022727 | 24023275 | LOC_Os03g43060 |
| Inversion | Chr12 | 24521150 | 24521299 | LOC_Os12g39700 |
| Inversion | Chr5 | 24638997 | 24639191 | |
| Inversion | Chr2 | 24687304 | 24687886 | LOC_Os02g40720 |
| Inversion | Chr2 | 24750714 | 24750858 | LOC_Os02g40840 |
| Inversion | Chr12 | 25358511 | 25358828 | LOC_Os12g40950 |
| Inversion | Chr6 | 25385692 | 25386094 | |
| Inversion | Chr2 | 25424030 | 25424426 | LOC_Os02g42280 |
| Inversion | Chr7 | 25659266 | 25659447 | LOC_Os07g42834 |
| Inversion | Chr7 | 25712487 | 25712612 | |
| Inversion | Chr1 | 25988547 | 25988749 | |
| Inversion | Chr5 | 26172813 | 26173159 | |
| Inversion | Chr12 | 26213432 | 26213711 | |
| Inversion | Chr7 | 26301468 | 26301918 | LOC_Os07g44000 |
| Inversion | Chr8 | 26515541 | 26515721 | |
| Inversion | Chr11 | 26706957 | 26707418 | |
| Inversion | Chr11 | 26723783 | 26724075 | 2 |
| Inversion | Chr3 | 26756738 | 26757287 | |
| Inversion | Chr6 | 26785600 | 26785779 | LOC_Os06g44380 |
| Inversion | Chr8 | 26987114 | 26987621 | |
| Inversion | Chr3 | 27526555 | 27526744 | LOC_Os03g48330 |
| Inversion | Chr3 | 27709226 | 27709365 | LOC_Os03g48600 |
| Inversion | Chr5 | 28443591 | 28443898 | LOC_Os05g49580 |
| Inversion | Chr6 | 28516933 | 28517176 | |
| Inversion | Chr1 | 28924396 | 28924532 | 2 |
| Inversion | Chr4 | 29208271 | 29208465 | LOC_Os04g48980 |
| Inversion | Chr3 | 29659055 | 29659229 | |
| Inversion | Chr4 | 29876530 | 29876987 | |
| Inversion | Chr2 | 29948145 | 29948430 | LOC_Os02g48980 |
| Inversion | Chr6 | 30158123 | 30158279 | |
| Inversion | Chr3 | 30269940 | 30270102 | LOC_Os03g52794 |
| Inversion | Chr3 | 31021183 | 31021617 | LOC_Os03g54114 |
| Inversion | Chr2 | 31225927 | 31226030 | |
| Inversion | Chr3 | 31400658 | 31401011 | LOC_Os03g55180 |
| Inversion | Chr1 | 32119784 | 32119982 | |
| Inversion | Chr4 | 32254473 | 32254793 | LOC_Os04g54140 |
| Inversion | Chr2 | 32284815 | 32285028 | LOC_Os02g52800 |
| Inversion | Chr4 | 32798715 | 32798851 | |
| Inversion | Chr2 | 33386986 | 33387165 | LOC_Os02g54510 |
| Inversion | Chr4 | 33441828 | 33442050 | |
| Inversion | Chr1 | 34936798 | 34937054 | |
| Inversion | Chr4 | 34973316 | 34973735 | |
| Inversion | Chr4 | 35107746 | 35108042 | |
| Inversion | Chr2 | 35498477 | 35498662 | LOC_Os02g57960 |
| Inversion | Chr1 | 36965842 | 36966095 | |
| Inversion | Chr1 | 37094407 | 37094676 | |
| Inversion | Chr1 | 39564883 | 39565079 | LOC_Os01g68050 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 3773762 | Chr2 | 15357418 | LOC_Os06g07780 |
| Translocation | Chr4 | 7546307 | Chr1 | 2551414 | 2 |
| Translocation | Chr10 | 8097515 | Chr1 | 16931702 | |
| Translocation | Chr7 | 22644370 | Chr6 | 3770448 | 2 |
