Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1686-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1686-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 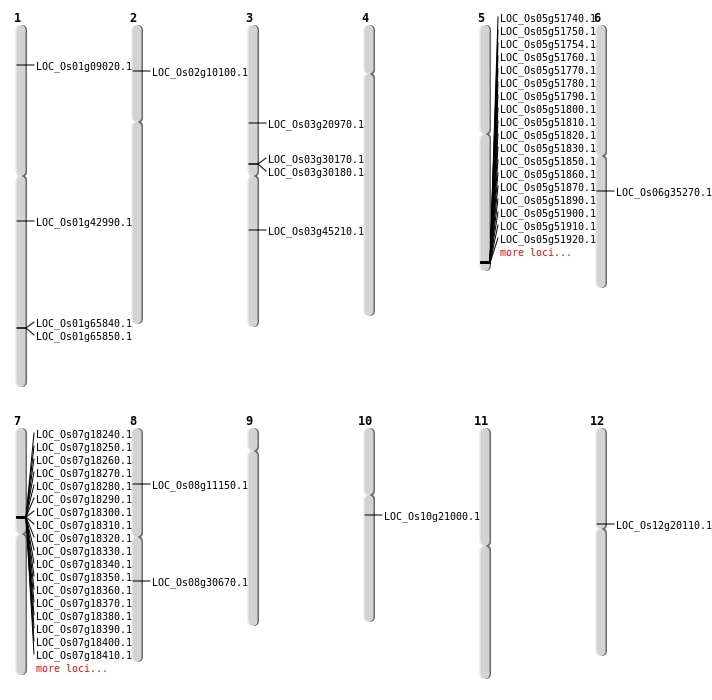
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11113743 | T-C | Homo | ||
| SBS | Chr1 | 24830824 | G-A | HET | ||
| SBS | Chr10 | 13007890 | G-T | HET | ||
| SBS | Chr10 | 15157922 | G-T | HET | ||
| SBS | Chr11 | 24846925 | T-A | HET | ||
| SBS | Chr11 | 641217 | C-T | HET | ||
| SBS | Chr12 | 11694756 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g20110 |
| SBS | Chr12 | 13335636 | T-A | HET | ||
| SBS | Chr12 | 21567611 | C-T | HET | ||
| SBS | Chr12 | 24044074 | G-A | Homo | ||
| SBS | Chr2 | 15923141 | T-A | HET | ||
| SBS | Chr2 | 28969382 | C-A | Homo | ||
| SBS | Chr2 | 30918847 | T-C | Homo | ||
| SBS | Chr2 | 32437411 | C-T | Homo | ||
| SBS | Chr2 | 5604034 | G-A | HET | ||
| SBS | Chr3 | 11897644 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g20970 |
| SBS | Chr4 | 10416415 | G-C | HET | ||
| SBS | Chr4 | 14319894 | A-T | HET | ||
| SBS | Chr4 | 23746182 | C-T | HET | ||
| SBS | Chr4 | 31939684 | C-T | HET | ||
| SBS | Chr4 | 33997299 | G-T | HET | ||
| SBS | Chr5 | 21929367 | A-T | Homo | ||
| SBS | Chr5 | 21929368 | A-T | Homo | ||
| SBS | Chr5 | 21929379 | C-T | Homo | ||
| SBS | Chr5 | 23707047 | G-T | Homo | ||
| SBS | Chr6 | 15041005 | C-A | Homo | ||
| SBS | Chr6 | 20564015 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g35270 |
| SBS | Chr7 | 11155021 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g18840 |
| SBS | Chr7 | 12986790 | G-C | HET | ||
| SBS | Chr7 | 13632673 | C-T | HET | ||
| SBS | Chr7 | 3852334 | T-G | HET | ||
| SBS | Chr7 | 794170 | C-A | HET | ||
| SBS | Chr8 | 2629601 | A-T | Homo | ||
| SBS | Chr8 | 28220186 | G-T | HET | ||
| SBS | Chr8 | 28220187 | C-T | HET | ||
| SBS | Chr8 | 6551618 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11150 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 762571 | 762572 | 1 | |
| Deletion | Chr11 | 1139840 | 1139844 | 4 | |
| Deletion | Chr2 | 1276435 | 1276437 | 2 | |
| Deletion | Chr3 | 2107204 | 2107209 | 5 | |
| Deletion | Chr1 | 3029062 | 3029067 | 5 | |
| Deletion | Chr1 | 3951643 | 3951645 | 2 | |
| Deletion | Chr10 | 4782290 | 4782298 | 8 | |
| Deletion | Chr3 | 7943427 | 7943428 | 1 | |
| Deletion | Chr1 | 8502717 | 8502745 | 28 | |
| Deletion | Chr10 | 10640033 | 10640043 | 10 | LOC_Os10g21000 |
| Deletion | Chr7 | 10811001 | 11140000 | 328999 | 53 |
| Deletion | Chr7 | 11401001 | 11548000 | 146999 | 23 |
| Deletion | Chr10 | 11434887 | 11434888 | 1 | |
| Deletion | Chr3 | 17126874 | 17126893 | 19 | |
| Deletion | Chr3 | 17220001 | 17224000 | 3999 | 2 |
| Deletion | Chr8 | 18906559 | 18906560 | 1 | LOC_Os08g30670 |
| Deletion | Chr5 | 21925318 | 21925321 | 3 | |
| Deletion | Chr3 | 23771650 | 23771654 | 4 | |
| Deletion | Chr11 | 27182187 | 27182197 | 10 | |
| Deletion | Chr5 | 29678001 | 29804000 | 125999 | 19 |
| Deletion | Chr5 | 29892001 | 29901000 | 8999 | 2 |
| Deletion | Chr2 | 31184821 | 31184823 | 2 |
Insertions: 5
Inversions: 10
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 2492686 | 2493096 | |
| Inversion | Chr1 | 4539256 | 4539397 | LOC_Os01g09020 |
| Inversion | Chr2 | 5262883 | 5263368 | LOC_Os02g10100 |
| Inversion | Chr4 | 11014444 | 11341741 | |
| Inversion | Chr4 | 16522544 | 16522747 | |
| Inversion | Chr6 | 17191751 | 17192059 | |
| Inversion | Chr1 | 24487615 | 24528653 | LOC_Os01g42990 |
| Inversion | Chr1 | 24487616 | 24528655 | LOC_Os01g42990 |
| Inversion | Chr1 | 38228430 | 38237042 | 2 |
| Inversion | Chr1 | 38228435 | 38237049 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 13220780 | Chr5 | 29713108 | |
| Translocation | Chr5 | 23219934 | Chr3 | 25528108 | 2 |
