Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1703-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1703-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 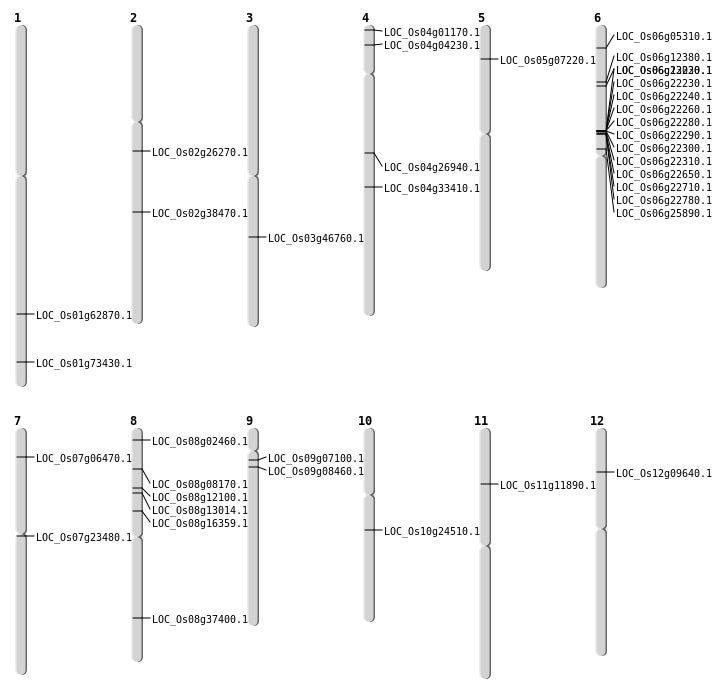
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28056993 | G-A | HET | ||
| SBS | Chr1 | 28056994 | T-A | HET | ||
| SBS | Chr10 | 1895170 | G-A | Homo | ||
| SBS | Chr10 | 19840084 | T-C | Homo | ||
| SBS | Chr10 | 5392997 | A-G | Homo | ||
| SBS | Chr10 | 5393004 | A-T | Homo | ||
| SBS | Chr11 | 22961382 | C-A | HET | ||
| SBS | Chr12 | 25604127 | G-A | Homo | ||
| SBS | Chr2 | 1057536 | A-T | HET | ||
| SBS | Chr2 | 11741964 | C-T | Homo | ||
| SBS | Chr3 | 13864560 | G-A | HET | ||
| SBS | Chr3 | 26452293 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46760 |
| SBS | Chr3 | 5021303 | A-G | HET | ||
| SBS | Chr3 | 5848050 | T-A | HET | ||
| SBS | Chr3 | 7321207 | A-G | HET | ||
| SBS | Chr4 | 1971643 | C-T | Homo | ||
| SBS | Chr4 | 1971646 | C-G | Homo | ||
| SBS | Chr4 | 1971650 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g04230 |
| SBS | Chr4 | 6959755 | C-T | Homo | ||
| SBS | Chr6 | 12440578 | A-T | Homo | ||
| SBS | Chr6 | 14891183 | C-T | Homo | ||
| SBS | Chr6 | 26320541 | G-A | Homo | ||
| SBS | Chr6 | 6712706 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g12380 |
| SBS | Chr7 | 1978566 | A-G | HET | ||
| SBS | Chr7 | 22710315 | G-A | HET | ||
| SBS | Chr7 | 27808674 | G-T | HET | ||
| SBS | Chr7 | 596157 | G-A | HET | ||
| SBS | Chr8 | 15910403 | T-C | HET | ||
| SBS | Chr8 | 23664592 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g37400 |
| SBS | Chr8 | 23950858 | G-A | HET | ||
| SBS | Chr9 | 4413723 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g08460 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 84604 | 84958 | 354 | |
| Deletion | Chr4 | 598620 | 598623 | 3 | |
| Deletion | Chr9 | 787958 | 787959 | 1 | |
| Deletion | Chr1 | 1259504 | 1259923 | 419 | |
| Deletion | Chr3 | 1358513 | 1358514 | 1 | |
| Deletion | Chr4 | 1614704 | 1614708 | 4 | |
| Deletion | Chr2 | 11738340 | 11738341 | 1 | |
| Deletion | Chr11 | 12087648 | 12087651 | 3 | |
| Deletion | Chr10 | 12583910 | 12583911 | 1 | LOC_Os10g24510 |
| Deletion | Chr6 | 12887001 | 12955000 | 67999 | 8 |
| Deletion | Chr8 | 13782176 | 13782177 | 1 | |
| Deletion | Chr6 | 15138344 | 15138345 | 1 | LOC_Os06g25890 |
| Deletion | Chr8 | 17051091 | 17051092 | 1 | |
| Deletion | Chr8 | 18763199 | 18763200 | 1 | |
| Deletion | Chr4 | 20234533 | 20234535 | 2 | LOC_Os04g33410 |
| Deletion | Chr3 | 27725836 | 27725842 | 6 | |
| Deletion | Chr1 | 42553508 | 42553512 | 4 | LOC_Os01g73430 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 15983382 | 15983459 | 78 | |
| Insertion | Chr11 | 12115576 | 12115576 | 1 | |
| Insertion | Chr11 | 2729463 | 2729526 | 64 | |
| Insertion | Chr11 | 27370142 | 27370142 | 1 | |
| Insertion | Chr12 | 23760671 | 23760742 | 72 | |
| Insertion | Chr4 | 129110 | 129115 | 6 | LOC_Os04g01170 |
| Insertion | Chr5 | 12568234 | 12568234 | 1 |
Inversions: 20
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 984767 | 985024 | LOC_Os08g02460 |
| Inversion | Chr6 | 2387897 | 2388180 | LOC_Os06g05310 |
| Inversion | Chr7 | 3128357 | 3128636 | LOC_Os07g06470 |
| Inversion | Chr5 | 3816332 | 3816587 | LOC_Os05g07220 |
| Inversion | Chr12 | 5079759 | 5167034 | LOC_Os12g09640 |
| Inversion | Chr12 | 5079766 | 5167048 | LOC_Os12g09640 |
| Inversion | Chr6 | 7135270 | 7135512 | LOC_Os06g13030 |
| Inversion | Chr12 | 8029020 | 8029250 | |
| Inversion | Chr8 | 9092449 | 9092568 | |
| Inversion | Chr6 | 11360675 | 11416581 | |
| Inversion | Chr6 | 12886846 | 13163291 | 2 |
| Inversion | Chr6 | 12954575 | 13210202 | 2 |
| Inversion | Chr6 | 13163300 | 13263150 | 2 |
| Inversion | Chr8 | 17072251 | 17072550 | |
| Inversion | Chr9 | 19471648 | 19471831 | |
| Inversion | Chr2 | 20875995 | 20876790 | |
| Inversion | Chr7 | 21213183 | 21213301 | |
| Inversion | Chr2 | 23259485 | 23259713 | LOC_Os02g38470 |
| Inversion | Chr12 | 23986652 | 24332913 | |
| Inversion | Chr7 | 27672568 | 27672816 |
Translocations: 18
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2785663 | Chr8 | 4650014 | 2 |
| Translocation | Chr9 | 3446706 | Chr8 | 7125803 | 2 |
| Translocation | Chr11 | 4231113 | Chr1 | 31989862 | |
| Translocation | Chr8 | 4650007 | Chr2 | 21197283 | LOC_Os08g08170 |
| Translocation | Chr11 | 5009699 | Chr1 | 36411664 | 2 |
| Translocation | Chr11 | 6602442 | Chr8 | 2455439 | LOC_Os11g11890 |
| Translocation | Chr11 | 13220824 | Chr1 | 40950394 | |
| Translocation | Chr7 | 13266463 | Chr3 | 14801962 | LOC_Os07g23480 |
| Translocation | Chr10 | 13764672 | Chr2 | 29617581 | |
| Translocation | Chr4 | 15937086 | Chr2 | 15425627 | 2 |
| Translocation | Chr12 | 16959412 | Chr8 | 9984465 | 2 |
| Translocation | Chr11 | 17701376 | Chr8 | 24046280 | |
| Translocation | Chr6 | 19630066 | Chr3 | 24413583 | |
| Translocation | Chr12 | 21526175 | Chr8 | 7731028 | 2 |
| Translocation | Chr7 | 22194697 | Chr3 | 17909148 | |
| Translocation | Chr12 | 22237226 | Chr1 | 13535625 | |
| Translocation | Chr6 | 27084503 | Chr1 | 27776529 | |
| Translocation | Chr12 | 27107844 | Chr7 | 22632770 |
