Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1734-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1734-S Alignment File |
| Seed Availability | No |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 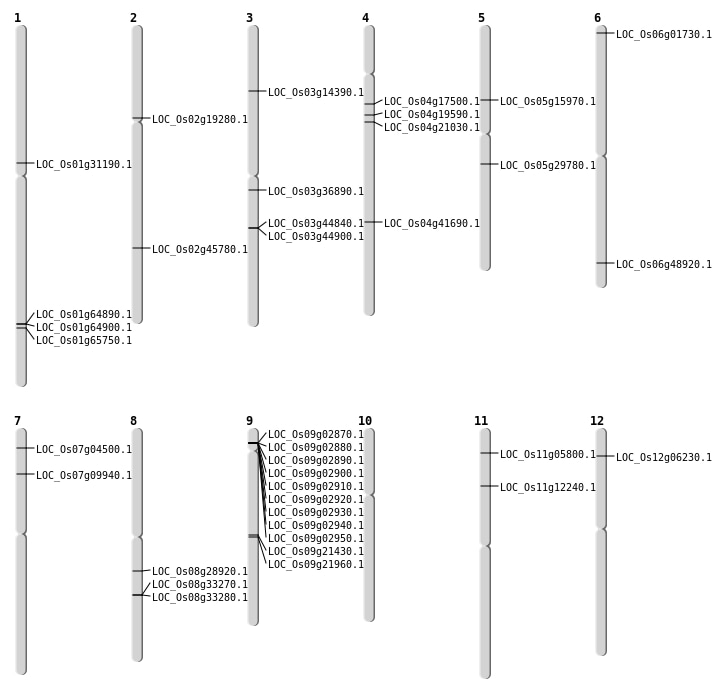
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 36075788 | G-A | HET | ||
| SBS | Chr1 | 39845783 | C-G | HET | ||
| SBS | Chr10 | 21982380 | C-G | HET | ||
| SBS | Chr10 | 22905633 | G-T | HET | ||
| SBS | Chr2 | 1697287 | T-A | HET | ||
| SBS | Chr2 | 27272004 | C-A | HET | ||
| SBS | Chr3 | 20364070 | G-A | HET | ||
| SBS | Chr4 | 12754778 | G-C | HET | ||
| SBS | Chr4 | 8712135 | G-C | HET | ||
| SBS | Chr5 | 11753304 | C-G | Homo | ||
| SBS | Chr5 | 20214090 | G-A | HET | ||
| SBS | Chr6 | 14548126 | T-C | HET | ||
| SBS | Chr6 | 17540443 | A-T | HET | ||
| SBS | Chr6 | 18214619 | T-A | HET | ||
| SBS | Chr6 | 3088930 | G-T | HET | ||
| SBS | Chr7 | 26760017 | G-A | Homo | ||
| SBS | Chr7 | 5293606 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g09940 |
| SBS | Chr7 | 5897496 | T-A | Homo | ||
| SBS | Chr8 | 2579479 | C-A | Homo | ||
| SBS | Chr9 | 1334519 | C-G | Homo | ||
| SBS | Chr9 | 17760291 | G-A | Homo |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1344001 | 1390000 | 45999 | 9 |
| Deletion | Chr8 | 4028096 | 4028097 | 1 | |
| Deletion | Chr5 | 9017197 | 9017209 | 12 | LOC_Os05g15970 |
| Deletion | Chr4 | 9549507 | 9549509 | 2 | |
| Deletion | Chr4 | 9576661 | 9576726 | 65 | LOC_Os04g17500 |
| Deletion | Chr12 | 10520727 | 10520728 | 1 | |
| Deletion | Chr4 | 11826572 | 11826573 | 1 | LOC_Os04g21030 |
| Deletion | Chr2 | 11964596 | 11964597 | 1 | |
| Deletion | Chr1 | 17056062 | 17056088 | 26 | LOC_Os01g31190 |
| Deletion | Chr8 | 20729001 | 20741000 | 11999 | 2 |
| Deletion | Chr4 | 23252001 | 23262000 | 9999 | |
| Deletion | Chr2 | 23921001 | 23940000 | 18999 | |
| Deletion | Chr4 | 24713489 | 24713490 | 1 | LOC_Os04g41690 |
| Deletion | Chr3 | 26905001 | 26916000 | 10999 | |
| Deletion | Chr2 | 27880120 | 27880125 | 5 | |
| Deletion | Chr6 | 29619020 | 29619021 | 1 | LOC_Os06g48920 |
| Deletion | Chr1 | 37667001 | 37678000 | 10999 | 2 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 41591047 | 41591047 | 1 | |
| Insertion | Chr11 | 25772785 | 25772787 | 3 | |
| Insertion | Chr12 | 11995653 | 11995653 | 1 | |
| Insertion | Chr12 | 15721069 | 15721070 | 2 | |
| Insertion | Chr12 | 3369689 | 3369690 | 2 | |
| Insertion | Chr3 | 12564474 | 12564475 | 2 | |
| Insertion | Chr3 | 16852251 | 16852252 | 2 | |
| Insertion | Chr4 | 10182469 | 10182469 | 1 | |
| Insertion | Chr4 | 10915131 | 10915131 | 1 | LOC_Os04g19590 |
| Insertion | Chr4 | 2313217 | 2313218 | 2 | |
| Insertion | Chr5 | 7664025 | 7664025 | 1 | |
| Insertion | Chr7 | 22554021 | 22554021 | 1 | |
| Insertion | Chr7 | 27766716 | 27766750 | 35 |
Inversions: 17
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1987098 | 1987413 | LOC_Os07g04500 |
| Inversion | Chr11 | 2682625 | 2682944 | 2 |
| Inversion | Chr5 | 6129687 | 6482207 | |
| Inversion | Chr5 | 6129699 | 6482219 | |
| Inversion | Chr11 | 6641847 | 6832561 | 2 |
| Inversion | Chr3 | 7827865 | 7828270 | 2 |
| Inversion | Chr2 | 12416310 | 12978543 | LOC_Os02g20980 |
| Inversion | Chr9 | 13297181 | 13297474 | LOC_Os09g21960 |
| Inversion | Chr12 | 14537244 | 14538633 | |
| Inversion | Chr12 | 14537412 | 14538940 | |
| Inversion | Chr8 | 17682322 | 17683531 | 2 |
| Inversion | Chr9 | 19432913 | 19433312 | |
| Inversion | Chr1 | 20105391 | 20106615 | |
| Inversion | Chr7 | 21840737 | 21840885 | |
| Inversion | Chr3 | 25299027 | 25299223 | LOC_Os03g44840 |
| Inversion | Chr4 | 27757786 | 27758046 | |
| Inversion | Chr2 | 27887143 | 27887290 | LOC_Os02g45780 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 428097 | Chr3 | 23730051 | LOC_Os06g01730 |
| Translocation | Chr7 | 2280346 | Chr4 | 20113981 | |
| Translocation | Chr9 | 2541645 | Chr6 | 21522627 | |
| Translocation | Chr12 | 2960173 | Chr2 | 11267535 | 2 |
| Translocation | Chr9 | 5669484 | Chr2 | 17526015 | |
| Translocation | Chr10 | 10540511 | Chr1 | 38170057 | 2 |
| Translocation | Chr9 | 12955377 | Chr1 | 33750971 | LOC_Os09g21430 |
| Translocation | Chr5 | 17232419 | Chr3 | 20467848 | 2 |
| Translocation | Chr10 | 20738872 | Chr3 | 25343032 | 2 |
| Translocation | Chr7 | 24489686 | Chr2 | 7072515 | |
| Translocation | Chr3 | 32459731 | Chr1 | 35462498 |
