Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1735-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1735-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 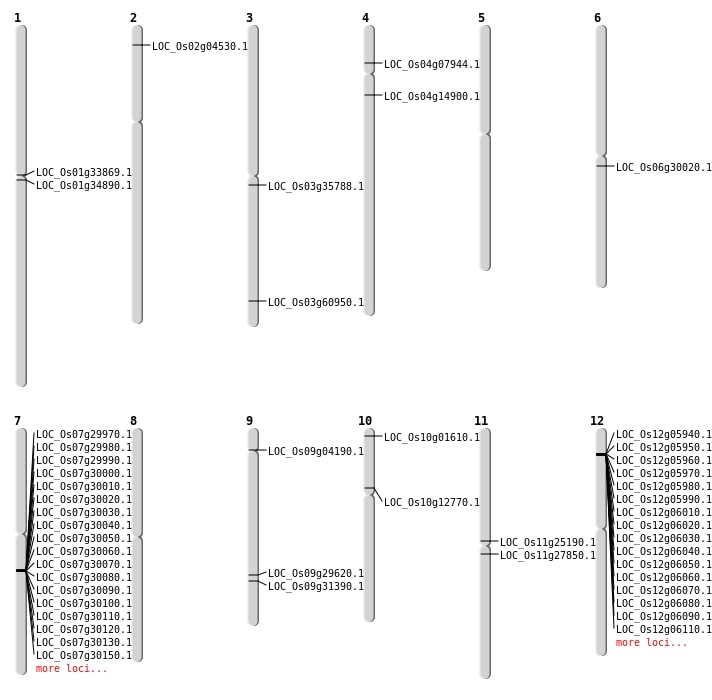
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27687661 | G-T | HET | ||
| SBS | Chr1 | 32043054 | A-G | HET | ||
| SBS | Chr1 | 7140538 | C-T | HET | ||
| SBS | Chr10 | 11013703 | C-T | Homo | ||
| SBS | Chr10 | 12531123 | C-T | Homo | ||
| SBS | Chr10 | 16316636 | A-G | HET | ||
| SBS | Chr10 | 17239467 | T-C | HET | ||
| SBS | Chr10 | 413270 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g01610 |
| SBS | Chr10 | 7130967 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g12770 |
| SBS | Chr11 | 21113191 | A-T | HET | ||
| SBS | Chr11 | 5646325 | T-A | HET | ||
| SBS | Chr11 | 7412217 | G-A | HET | ||
| SBS | Chr11 | 8520170 | C-T | HET | ||
| SBS | Chr12 | 15225537 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26140 |
| SBS | Chr12 | 19221222 | C-A | HET | ||
| SBS | Chr12 | 3995526 | A-G | Homo | ||
| SBS | Chr2 | 24157771 | G-A | HET | ||
| SBS | Chr2 | 35542152 | T-C | HET | ||
| SBS | Chr3 | 13112463 | T-C | HET | ||
| SBS | Chr3 | 13113200 | G-C | HET | ||
| SBS | Chr3 | 15523904 | C-T | HET | ||
| SBS | Chr3 | 19640645 | T-C | HET | ||
| SBS | Chr3 | 19834066 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g35788 |
| SBS | Chr3 | 29750257 | C-T | Homo | ||
| SBS | Chr3 | 7034572 | G-A | Homo | ||
| SBS | Chr4 | 29314722 | C-G | HET | ||
| SBS | Chr4 | 29361230 | T-A | HET | ||
| SBS | Chr4 | 3865011 | C-A | HET | ||
| SBS | Chr4 | 4235414 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07944 |
| SBS | Chr4 | 8385329 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14900 |
| SBS | Chr6 | 1107683 | G-A | HET | ||
| SBS | Chr6 | 17279524 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g30020 |
| SBS | Chr6 | 17364138 | G-A | Homo | ||
| SBS | Chr6 | 1764994 | T-C | HET | ||
| SBS | Chr6 | 22966454 | T-C | HET | ||
| SBS | Chr6 | 30588023 | C-T | HET | ||
| SBS | Chr7 | 21153658 | C-T | HET | ||
| SBS | Chr8 | 6685630 | T-C | HET | ||
| SBS | Chr9 | 2562488 | T-A | HET |
Deletions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2206521 | 2206522 | 1 | LOC_Os09g04190 |
| Deletion | Chr12 | 2739001 | 2897000 | 157999 | 20 |
| Deletion | Chr12 | 3274288 | 3274303 | 15 | |
| Deletion | Chr12 | 4141398 | 4141399 | 1 | |
| Deletion | Chr2 | 10194678 | 10194679 | 1 | |
| Deletion | Chr7 | 17171326 | 17171329 | 3 | |
| Deletion | Chr7 | 17647001 | 17847000 | 199999 | 25 |
| Deletion | Chr8 | 19206441 | 19206452 | 11 | |
| Deletion | Chr12 | 22162589 | 22162595 | 6 | |
| Deletion | Chr4 | 23112158 | 23112162 | 4 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 24157573 | 24157573 | 1 | |
| Insertion | Chr8 | 20836443 | 20836444 | 2 |
Inversions: 13
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 2015503 | 2015874 | LOC_Os02g04530 |
| Inversion | Chr12 | 2669995 | 2738612 | 2 |
| Inversion | Chr6 | 6624712 | 7046536 | |
| Inversion | Chr6 | 6624729 | 7046547 | |
| Inversion | Chr9 | 15563937 | 15564375 | |
| Inversion | Chr4 | 16990382 | 16990616 | |
| Inversion | Chr9 | 18012180 | 18012435 | LOC_Os09g29620 |
| Inversion | Chr1 | 18496345 | 18789915 | |
| Inversion | Chr9 | 18871802 | 18872107 | LOC_Os09g31390 |
| Inversion | Chr1 | 19266542 | 19342635 | LOC_Os01g34890 |
| Inversion | Chr7 | 21517681 | 22275177 | |
| Inversion | Chr1 | 29678525 | 29678648 | |
| Inversion | Chr3 | 34634233 | 34634529 | LOC_Os03g60950 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2670005 | Chr11 | 16030297 | 2 |
| Translocation | Chr12 | 2896320 | Chr11 | 16030105 | 2 |
| Translocation | Chr11 | 14356146 | Chr7 | 21168221 | 2 |
| Translocation | Chr11 | 14356351 | Chr7 | 21693128 | 2 |
| Translocation | Chr7 | 18622180 | Chr1 | 18638037 | 2 |
| Translocation | Chr12 | 20710693 | Chr10 | 6314387 | LOC_Os12g34160 |
| Translocation | Chr7 | 21680216 | Chr4 | 26240978 | |
| Translocation | Chr7 | 21693122 | Chr4 | 26241053 | LOC_Os07g36280 |
