Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1737-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1737-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 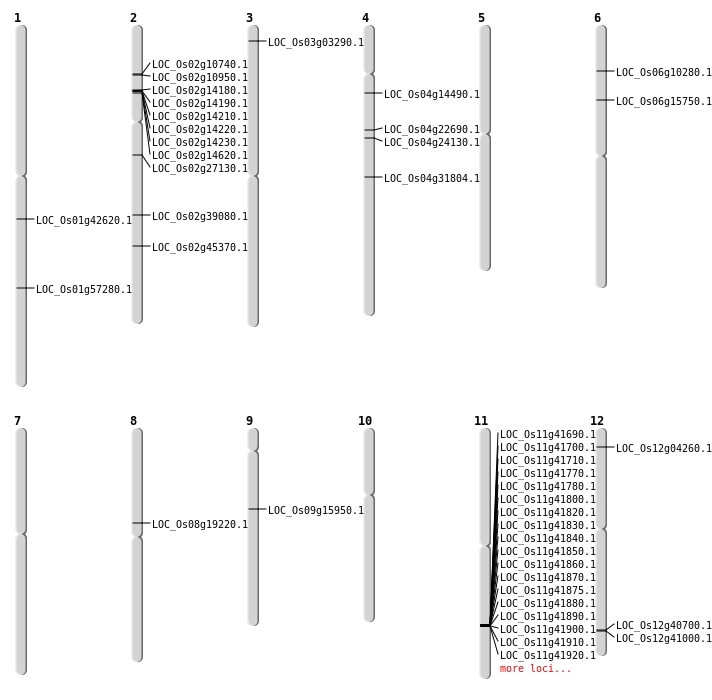
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24243844 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g42620 |
| SBS | Chr1 | 28253484 | C-T | HET | ||
| SBS | Chr1 | 33099868 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g57280 |
| SBS | Chr1 | 5665142 | C-T | HET | ||
| SBS | Chr10 | 12147670 | T-C | HET | ||
| SBS | Chr11 | 14886997 | C-T | HET | ||
| SBS | Chr11 | 14886998 | C-T | HET | ||
| SBS | Chr11 | 22222037 | G-T | HET | ||
| SBS | Chr11 | 3546874 | T-A | Homo | ||
| SBS | Chr11 | 3546876 | T-A | Homo | ||
| SBS | Chr11 | 3546879 | T-A | Homo | ||
| SBS | Chr12 | 15098290 | C-T | HET | ||
| SBS | Chr12 | 1803336 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04260 |
| SBS | Chr12 | 1803337 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04260 |
| SBS | Chr12 | 23096761 | A-T | Homo | ||
| SBS | Chr12 | 25376031 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41000 |
| SBS | Chr2 | 10401778 | T-A | Homo | ||
| SBS | Chr2 | 13884176 | G-A | Homo | ||
| SBS | Chr2 | 27580454 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g45370 |
| SBS | Chr2 | 4213916 | A-G | HET | ||
| SBS | Chr2 | 7566517 | C-A | HET | ||
| SBS | Chr2 | 928490 | A-G | Homo | ||
| SBS | Chr3 | 2655683 | C-G | HET | ||
| SBS | Chr3 | 28268515 | G-A | HET | ||
| SBS | Chr3 | 7456805 | C-T | HET | ||
| SBS | Chr4 | 20146712 | C-T | HET | ||
| SBS | Chr4 | 8104219 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g14490 |
| SBS | Chr5 | 20559590 | G-T | HET | ||
| SBS | Chr5 | 3987987 | A-C | Homo | ||
| SBS | Chr6 | 12189493 | C-T | HET | ||
| SBS | Chr6 | 29131839 | G-A | HET | ||
| SBS | Chr6 | 6708126 | A-T | HET | ||
| SBS | Chr7 | 11208368 | G-A | Homo | ||
| SBS | Chr7 | 25303821 | G-A | HET | ||
| SBS | Chr8 | 11774021 | C-T | Homo | ||
| SBS | Chr8 | 5194750 | G-A | Homo | ||
| SBS | Chr9 | 13835888 | G-A | HET | ||
| SBS | Chr9 | 15733328 | G-A | HET | ||
| SBS | Chr9 | 4477862 | C-T | HET | ||
| SBS | Chr9 | 9744082 | G-T | HET | STOP_GAINED | LOC_Os09g15950 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1157217 | 1157218 | 1 | |
| Deletion | Chr7 | 2095462 | 2095463 | 1 | |
| Deletion | Chr8 | 2503356 | 2503358 | 2 | |
| Deletion | Chr10 | 4089629 | 4089755 | 126 | |
| Deletion | Chr6 | 5285847 | 5285859 | 12 | LOC_Os06g10280 |
| Deletion | Chr5 | 7263194 | 7263195 | 1 | |
| Deletion | Chr2 | 7764001 | 7798000 | 33999 | 5 |
| Deletion | Chr2 | 7922833 | 7922834 | 1 | |
| Deletion | Chr7 | 10222991 | 10222992 | 1 | |
| Deletion | Chr4 | 11507508 | 11507512 | 4 | |
| Deletion | Chr10 | 11619732 | 11619733 | 1 | |
| Deletion | Chr2 | 15964478 | 15964489 | 11 | LOC_Os02g27130 |
| Deletion | Chr4 | 16954093 | 16954102 | 9 | |
| Deletion | Chr3 | 17724083 | 17724090 | 7 | |
| Deletion | Chr7 | 19671630 | 19671632 | 2 | |
| Deletion | Chr2 | 21417901 | 21417902 | 1 | |
| Deletion | Chr11 | 25032001 | 25049000 | 16999 | 3 |
| Deletion | Chr11 | 25078001 | 25272000 | 193999 | 23 |
| Deletion | Chr11 | 25416001 | 25766000 | 349999 | 59 |
| Deletion | Chr6 | 26744083 | 26744084 | 1 | |
| Deletion | Chr7 | 29530802 | 29530820 | 18 | |
| Deletion | Chr1 | 31303814 | 31303815 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 29019533 | 29019608 | 76 | |
| Insertion | Chr10 | 15399535 | 15399611 | 77 | |
| Insertion | Chr11 | 22767910 | 22767910 | 1 |
Inversions: 10
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5663819 | 5818105 | 2 |
| Inversion | Chr2 | 5663826 | 5818113 | 2 |
| Inversion | Chr2 | 7763986 | 8070321 | 2 |
| Inversion | Chr2 | 7798587 | 8070337 | 2 |
| Inversion | Chr6 | 8937569 | 9029717 | LOC_Os06g15750 |
| Inversion | Chr6 | 8937584 | 9029726 | LOC_Os06g15750 |
| Inversion | Chr4 | 12858536 | 13797159 | 2 |
| Inversion | Chr4 | 12858815 | 13797190 | 2 |
| Inversion | Chr4 | 19044779 | 19044884 | LOC_Os04g31804 |
| Inversion | Chr2 | 23606543 | 23606726 | LOC_Os02g39080 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 1411200 | Chr2 | 16564865 | LOC_Os03g03290 |
| Translocation | Chr3 | 1411236 | Chr2 | 16571156 | LOC_Os03g03290 |
| Translocation | Chr12 | 25194111 | Chr8 | 11496200 | 2 |
