Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1746-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1746-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 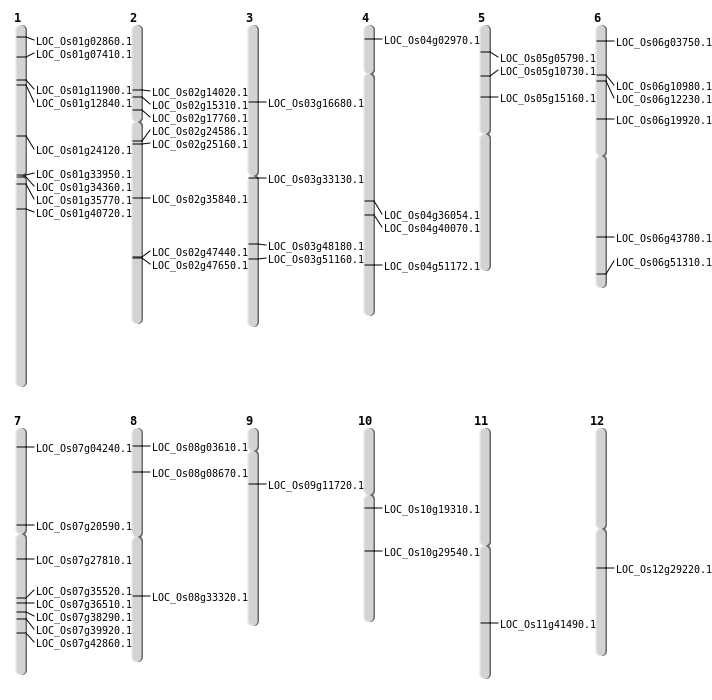
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18948228 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34360 |
| SBS | Chr1 | 22000475 | C-A | Homo | ||
| SBS | Chr1 | 30654252 | T-C | Homo | ||
| SBS | Chr10 | 1269640 | T-C | HET | ||
| SBS | Chr10 | 1318857 | T-G | HET | ||
| SBS | Chr10 | 4505724 | T-G | HET | ||
| SBS | Chr10 | 9557998 | C-T | HET | ||
| SBS | Chr11 | 20511938 | C-G | HET | ||
| SBS | Chr12 | 11627402 | C-G | HET | ||
| SBS | Chr12 | 1751157 | T-G | Homo | ||
| SBS | Chr12 | 7625510 | T-C | HET | ||
| SBS | Chr2 | 11291697 | C-T | HET | ||
| SBS | Chr2 | 14045315 | A-G | HET | ||
| SBS | Chr2 | 14459775 | G-A | HET | ||
| SBS | Chr2 | 294985 | A-T | Homo | ||
| SBS | Chr3 | 13197098 | G-A | Homo | ||
| SBS | Chr3 | 25397205 | G-A | Homo | ||
| SBS | Chr4 | 23859513 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g40070 |
| SBS | Chr4 | 25605032 | G-A | HET | ||
| SBS | Chr4 | 5418157 | G-A | HET | ||
| SBS | Chr5 | 17723987 | C-G | HET | ||
| SBS | Chr6 | 16110534 | A-G | HET | ||
| SBS | Chr6 | 8257809 | A-G | Homo | ||
| SBS | Chr7 | 5490354 | C-T | Homo | ||
| SBS | Chr8 | 24424482 | T-C | Homo | ||
| SBS | Chr8 | 442577 | G-T | HET | ||
| SBS | Chr8 | 5018954 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08670 |
| SBS | Chr9 | 21424037 | G-A | Homo |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2055218 | 2055219 | 1 | |
| Deletion | Chr8 | 3260257 | 3260268 | 11 | |
| Deletion | Chr12 | 4019824 | 4019826 | 2 | |
| Deletion | Chr1 | 4473716 | 4473717 | 1 | |
| Deletion | Chr10 | 5180144 | 5180147 | 3 | |
| Deletion | Chr4 | 7898054 | 7898059 | 5 | |
| Deletion | Chr6 | 11647912 | 11647913 | 1 | |
| Deletion | Chr9 | 14141269 | 14141272 | 3 | |
| Deletion | Chr2 | 14247782 | 14247783 | 1 | LOC_Os02g24586 |
| Deletion | Chr10 | 15380928 | 15380936 | 8 | |
| Deletion | Chr3 | 18954935 | 18954938 | 3 | LOC_Os03g33130 |
| Deletion | Chr12 | 20907697 | 20907703 | 6 | |
| Deletion | Chr12 | 21105677 | 21105679 | 2 | |
| Deletion | Chr11 | 24802358 | 24802359 | 1 | |
| Deletion | Chr1 | 25494351 | 25494352 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 15571121 | 15571122 | 2 | |
| Insertion | Chr10 | 5025186 | 5025186 | 1 | |
| Insertion | Chr6 | 4568915 | 4568980 | 66 |
Inversions: 91
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 370779 | 371119 | |
| Inversion | Chr5 | 985855 | 986120 | |
| Inversion | Chr1 | 1002373 | 1002572 | 2 |
| Inversion | Chr4 | 1201556 | 1201772 | LOC_Os04g02970 |
| Inversion | Chr3 | 1667038 | 1667217 | |
| Inversion | Chr8 | 1688204 | 1688543 | LOC_Os08g03610 |
| Inversion | Chr12 | 1764713 | 1764930 | |
| Inversion | Chr3 | 1770982 | 1771136 | |
| Inversion | Chr7 | 1862730 | 1862967 | LOC_Os07g04240 |
| Inversion | Chr1 | 2045100 | 2045326 | |
| Inversion | Chr5 | 2885517 | 2885816 | LOC_Os05g05790 |
| Inversion | Chr2 | 3302893 | 3303155 | LOC_Os02g06584 |
| Inversion | Chr9 | 3358350 | 3358461 | |
| Inversion | Chr11 | 3501431 | 3501905 | |
| Inversion | Chr1 | 3512396 | 3512580 | LOC_Os01g07410 |
| Inversion | Chr3 | 3842196 | 3842325 | |
| Inversion | Chr1 | 4526872 | 4527072 | |
| Inversion | Chr6 | 5734302 | 5734553 | LOC_Os06g10980 |
| Inversion | Chr5 | 5910611 | 5910755 | LOC_Os05g10730 |
| Inversion | Chr6 | 6064816 | 6064972 | |
| Inversion | Chr1 | 6449534 | 6449739 | LOC_Os01g11900 |
| Inversion | Chr9 | 6539625 | 6539782 | LOC_Os09g11720 |
| Inversion | Chr6 | 6570671 | 6570970 | 2 |
| Inversion | Chr7 | 6992353 | 6992780 | |
| Inversion | Chr1 | 7105142 | 7105555 | LOC_Os01g12840 |
| Inversion | Chr2 | 7654663 | 7654940 | LOC_Os02g14020 |
| Inversion | Chr2 | 8440486 | 8440721 | |
| Inversion | Chr2 | 8554102 | 8554209 | LOC_Os02g15310 |
| Inversion | Chr5 | 8602211 | 8602602 | LOC_Os05g15160 |
| Inversion | Chr3 | 9217096 | 9217431 | LOC_Os03g16680 |
| Inversion | Chr2 | 10263256 | 10263586 | LOC_Os02g17760 |
| Inversion | Chr2 | 10359107 | 10359401 | |
| Inversion | Chr10 | 10545128 | 10545395 | |
| Inversion | Chr4 | 11342552 | 11342908 | |
| Inversion | Chr6 | 11398099 | 11398273 | LOC_Os06g19920 |
| Inversion | Chr7 | 11903183 | 11903537 | 2 |
| Inversion | Chr11 | 12047036 | 12047260 | |
| Inversion | Chr4 | 12747791 | 12747997 | |
| Inversion | Chr3 | 13003211 | 13003820 | |
| Inversion | Chr1 | 13175841 | 13175957 | |
| Inversion | Chr1 | 13595912 | 13596287 | LOC_Os01g24120 |
| Inversion | Chr11 | 13888254 | 13888465 | |
| Inversion | Chr2 | 14629954 | 14630070 | LOC_Os02g25160 |
| Inversion | Chr10 | 15350967 | 15351085 | LOC_Os10g29540 |
| Inversion | Chr10 | 15371803 | 15372113 | |
| Inversion | Chr6 | 16202869 | 16203033 | |
| Inversion | Chr7 | 16229839 | 16230017 | LOC_Os07g27810 |
| Inversion | Chr7 | 16450799 | 16451131 | |
| Inversion | Chr10 | 16591244 | 16591418 | |
| Inversion | Chr6 | 16900476 | 16900667 | |
| Inversion | Chr12 | 17303309 | 17303699 | LOC_Os12g29220 |
| Inversion | Chr9 | 17348710 | 17349074 | |
| Inversion | Chr9 | 17564219 | 17564461 | |
| Inversion | Chr9 | 18125124 | 18125366 | |
| Inversion | Chr1 | 18672943 | 18673380 | LOC_Os01g33950 |
| Inversion | Chr10 | 19405895 | 19406050 | |
| Inversion | Chr5 | 19571537 | 19571845 | |
| Inversion | Chr3 | 19608029 | 19608165 | |
| Inversion | Chr1 | 19813944 | 19814266 | LOC_Os01g35770 |
| Inversion | Chr9 | 20193896 | 20194175 | |
| Inversion | Chr8 | 20776390 | 20776557 | LOC_Os08g33320 |
| Inversion | Chr7 | 21245810 | 21246210 | LOC_Os07g35520 |
| Inversion | Chr2 | 21529975 | 21530271 | LOC_Os02g35840 |
| Inversion | Chr5 | 21555128 | 21555322 | |
| Inversion | Chr8 | 21804771 | 21804984 | |
| Inversion | Chr7 | 21830618 | 21830748 | LOC_Os07g36510 |
| Inversion | Chr4 | 22008325 | 22008506 | LOC_Os04g36054 |
| Inversion | Chr7 | 22220742 | 22220986 | |
| Inversion | Chr7 | 23001127 | 23001268 | LOC_Os07g38290 |
| Inversion | Chr1 | 23006005 | 23006139 | LOC_Os01g40720 |
| Inversion | Chr11 | 23564170 | 23564310 | |
| Inversion | Chr7 | 23949412 | 23949642 | LOC_Os07g39920 |
| Inversion | Chr12 | 24014837 | 24015080 | |
| Inversion | Chr7 | 24730691 | 24731013 | |
| Inversion | Chr11 | 24888643 | 24889060 | LOC_Os11g41490 |
| Inversion | Chr2 | 25256543 | 25256872 | |
| Inversion | Chr1 | 25550643 | 25550988 | |
| Inversion | Chr5 | 25587365 | 25587793 | |
| Inversion | Chr7 | 25663296 | 25663557 | LOC_Os07g42860 |
| Inversion | Chr4 | 25849786 | 25850139 | |
| Inversion | Chr6 | 26355291 | 26355732 | 2 |
| Inversion | Chr5 | 26765297 | 26765545 | |
| Inversion | Chr3 | 27416184 | 27416299 | LOC_Os03g48180 |
| Inversion | Chr3 | 28515307 | 28515683 | |
| Inversion | Chr2 | 28969146 | 28969529 | LOC_Os02g47440 |
| Inversion | Chr2 | 29113648 | 29113996 | LOC_Os02g47650 |
| Inversion | Chr3 | 29277561 | 29277739 | LOC_Os03g51160 |
| Inversion | Chr4 | 30303060 | 30303312 | LOC_Os04g51172 |
| Inversion | Chr3 | 30621736 | 30621918 | |
| Inversion | Chr6 | 31071632 | 31071756 | LOC_Os06g51310 |
| Inversion | Chr1 | 40468787 | 40469003 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1483366 | Chr1 | 3192780 | LOC_Os06g03750 |
| Translocation | Chr12 | 4820680 | Chr6 | 4274541 | |
| Translocation | Chr10 | 9840475 | Chr9 | 665643 | LOC_Os10g19310 |
| Translocation | Chr11 | 10814760 | Chr8 | 22529967 | |
| Translocation | Chr8 | 16259496 | Chr4 | 19019713 | |
| Translocation | Chr11 | 19832414 | Chr10 | 10710616 | |
| Translocation | Chr6 | 21950535 | Chr1 | 13413514 |
