Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1761-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1761-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 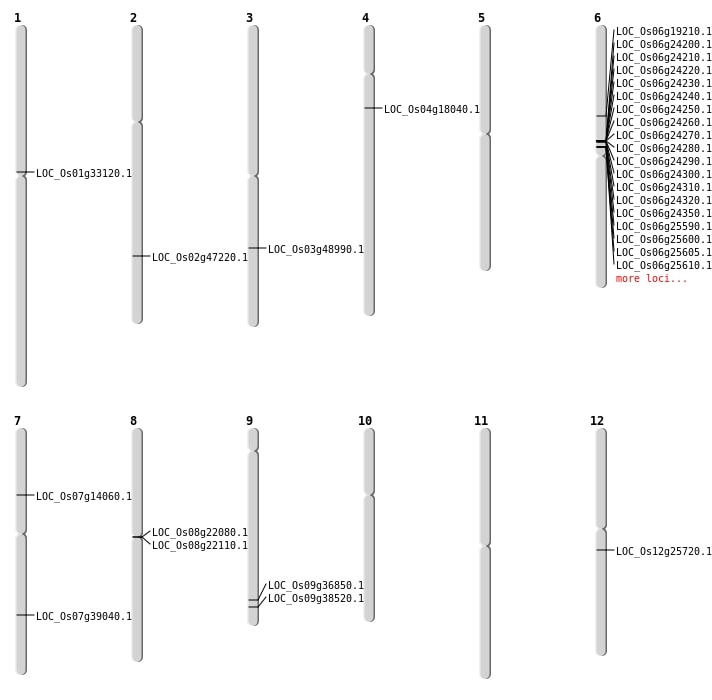
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20282606 | T-A | Homo | ||
| SBS | Chr1 | 20938947 | G-T | Homo | ||
| SBS | Chr1 | 28664344 | T-A | Homo | ||
| SBS | Chr1 | 33467625 | C-G | HET | ||
| SBS | Chr10 | 20074557 | A-T | Homo | ||
| SBS | Chr11 | 13390417 | A-G | Homo | ||
| SBS | Chr12 | 12612891 | G-T | HET | ||
| SBS | Chr12 | 13221775 | T-C | HET | ||
| SBS | Chr12 | 21569945 | G-A | HET | ||
| SBS | Chr2 | 11338698 | G-C | Homo | ||
| SBS | Chr2 | 19248589 | T-C | Homo | ||
| SBS | Chr2 | 28825581 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g47220 |
| SBS | Chr3 | 13559330 | C-T | Homo | ||
| SBS | Chr3 | 22619841 | T-G | HET | ||
| SBS | Chr3 | 24734812 | G-A | HET | ||
| SBS | Chr3 | 27903072 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g48990 |
| SBS | Chr3 | 34439027 | C-T | HET | ||
| SBS | Chr3 | 34439033 | C-T | HET | ||
| SBS | Chr4 | 14572693 | T-C | HET | ||
| SBS | Chr5 | 1114546 | T-A | Homo | ||
| SBS | Chr5 | 3079607 | A-T | Homo | ||
| SBS | Chr6 | 19758230 | A-G | HET | ||
| SBS | Chr6 | 4337561 | G-A | HET | ||
| SBS | Chr7 | 1533594 | C-T | HET | ||
| SBS | Chr7 | 20923497 | C-T | Homo | ||
| SBS | Chr7 | 24204869 | T-G | HET | ||
| SBS | Chr7 | 25927428 | G-A | HET | ||
| SBS | Chr7 | 8020454 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14060 |
| SBS | Chr9 | 21252569 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g36850 |
| SBS | Chr9 | 21767780 | C-T | HET | ||
| SBS | Chr9 | 22161503 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38520 |
| SBS | Chr9 | 22161505 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38520 |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1802770 | 1802771 | 1 | |
| Deletion | Chr2 | 2633762 | 2633764 | 2 | |
| Deletion | Chr8 | 5337766 | 5337767 | 1 | |
| Deletion | Chr6 | 6141558 | 6141559 | 1 | |
| Deletion | Chr1 | 10798045 | 10798046 | 1 | |
| Deletion | Chr7 | 12182670 | 12182671 | 1 | |
| Deletion | Chr8 | 13333994 | 13334002 | 8 | LOC_Os08g22080 |
| Deletion | Chr8 | 13342478 | 13342484 | 6 | LOC_Os08g22110 |
| Deletion | Chr6 | 14174001 | 14188000 | 13999 | 4 |
| Deletion | Chr6 | 14190001 | 14255000 | 64999 | 10 |
| Deletion | Chr12 | 14916489 | 14916490 | 1 | LOC_Os12g25720 |
| Deletion | Chr6 | 14966001 | 15076000 | 109999 | 23 |
Insertions: 7
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5902871 | Chr3 | 24413754 | |
| Translocation | Chr4 | 9968190 | Chr1 | 18234570 | 2 |
| Translocation | Chr6 | 10932697 | Chr3 | 23077806 | LOC_Os06g19210 |
| Translocation | Chr6 | 10932794 | Chr3 | 23077707 | LOC_Os06g19210 |
| Translocation | Chr7 | 23408491 | Chr4 | 22035009 | LOC_Os07g39040 |
| Translocation | Chr8 | 24077240 | Chr2 | 25044700 |
