Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2080-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2080-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 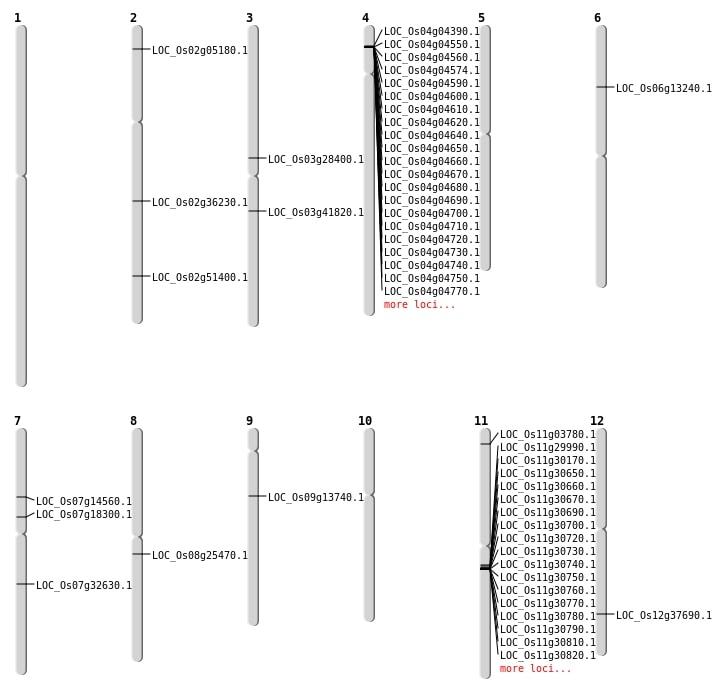
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13232029 | G-C | HET | ||
| SBS | Chr1 | 31378079 | T-G | HET | ||
| SBS | Chr1 | 32153541 | T-C | HET | ||
| SBS | Chr1 | 39528244 | A-C | HET | ||
| SBS | Chr1 | 5439574 | T-A | Homo | ||
| SBS | Chr11 | 24669461 | A-G | HET | ||
| SBS | Chr11 | 24942324 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g41580 |
| SBS | Chr11 | 7144791 | A-G | HET | ||
| SBS | Chr12 | 17675953 | G-A | Homo | ||
| SBS | Chr12 | 23202663 | C-T | HET | ||
| SBS | Chr12 | 27449942 | A-G | HET | ||
| SBS | Chr2 | 12695254 | C-T | HET | ||
| SBS | Chr2 | 14348740 | T-C | HET | ||
| SBS | Chr2 | 4527017 | G-A | HET | ||
| SBS | Chr2 | 4580644 | A-G | HET | ||
| SBS | Chr2 | 6769629 | C-A | HET | ||
| SBS | Chr3 | 16359854 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28400 |
| SBS | Chr3 | 7914700 | T-A | HET | ||
| SBS | Chr4 | 15916543 | G-C | HET | ||
| SBS | Chr4 | 16999862 | T-G | HET | ||
| SBS | Chr4 | 6322843 | G-A | HET | ||
| SBS | Chr4 | 6322844 | T-A | HET | ||
| SBS | Chr4 | 9384298 | G-A | HET | ||
| SBS | Chr4 | 9384299 | A-C | HET | ||
| SBS | Chr5 | 26216254 | T-C | HET | ||
| SBS | Chr5 | 470012 | G-A | Homo | ||
| SBS | Chr6 | 15671998 | T-C | HET | ||
| SBS | Chr6 | 20714236 | G-A | HET | ||
| SBS | Chr6 | 25216196 | T-C | HET | ||
| SBS | Chr7 | 10842949 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18300 |
| SBS | Chr7 | 13683830 | C-T | HET | ||
| SBS | Chr7 | 16525832 | T-C | Homo | ||
| SBS | Chr7 | 18712756 | T-A | Homo | ||
| SBS | Chr7 | 19420548 | C-T | Homo | ||
| SBS | Chr7 | 19708927 | A-T | Homo | ||
| SBS | Chr7 | 6966125 | C-A | HET | ||
| SBS | Chr7 | 8306434 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14560 |
| SBS | Chr9 | 11203382 | T-A | HET | ||
| SBS | Chr9 | 22158640 | C-A | HET | ||
| SBS | Chr9 | 3300834 | G-A | HET | ||
| SBS | Chr9 | 7057744 | A-G | Homo | ||
| SBS | Chr9 | 8046751 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13740 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1255020 | 1255021 | 1 | |
| Deletion | Chr9 | 2030486 | 2030490 | 4 | |
| Deletion | Chr4 | 2072001 | 2085000 | 12999 | LOC_Os04g04390 |
| Deletion | Chr4 | 2177001 | 2356000 | 178999 | 31 |
| Deletion | Chr4 | 2366001 | 2432000 | 65999 | 9 |
| Deletion | Chr4 | 2439001 | 2569000 | 129999 | 13 |
| Deletion | Chr4 | 2585001 | 2631000 | 45999 | 11 |
| Deletion | Chr4 | 2878001 | 2950000 | 71999 | 9 |
| Deletion | Chr5 | 3448903 | 3448913 | 10 | |
| Deletion | Chr2 | 10062448 | 10062449 | 1 | |
| Deletion | Chr11 | 13903289 | 13903291 | 2 | |
| Deletion | Chr4 | 16006377 | 16006378 | 1 | |
| Deletion | Chr9 | 16653800 | 16653806 | 6 | |
| Deletion | Chr11 | 17839001 | 18052000 | 212999 | 32 |
| Deletion | Chr7 | 19157418 | 19157439 | 21 | |
| Deletion | Chr7 | 19189721 | 19189722 | 1 | |
| Deletion | Chr7 | 19463606 | 19463625 | 19 | LOC_Os07g32630 |
| Deletion | Chr11 | 22062022 | 22062026 | 4 | |
| Deletion | Chr12 | 23132001 | 23146000 | 13999 | LOC_Os12g37690 |
| Deletion | Chr1 | 27428083 | 27428084 | 1 | |
| Deletion | Chr5 | 29119186 | 29119187 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 2798908 | 2798909 | 2 | |
| Insertion | Chr5 | 20461634 | 20461637 | 4 | |
| Insertion | Chr6 | 630575 | 630575 | 1 | |
| Insertion | Chr8 | 16212030 | 16212031 | 2 |
Inversions: 12
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 360892 | 1145481 | |
| Inversion | Chr11 | 1485259 | 1485398 | LOC_Os11g03780 |
| Inversion | Chr2 | 2474775 | 3391978 | LOC_Os02g05180 |
| Inversion | Chr2 | 2474780 | 3391980 | LOC_Os02g05180 |
| Inversion | Chr11 | 4455077 | 4455351 | |
| Inversion | Chr3 | 9151110 | 9151346 | |
| Inversion | Chr8 | 15502460 | 15502622 | LOC_Os08g25470 |
| Inversion | Chr11 | 17434917 | 18207873 | LOC_Os11g29990 |
| Inversion | Chr6 | 17874598 | 17874928 | |
| Inversion | Chr3 | 23221748 | 23222016 | LOC_Os03g41820 |
| Inversion | Chr2 | 31373358 | 31477337 | 2 |
| Inversion | Chr2 | 31373359 | 31477359 | 2 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 1148625 | Chr2 | 25364403 | |
| Translocation | Chr5 | 1148633 | Chr2 | 25364268 | |
| Translocation | Chr12 | 2363959 | Chr9 | 21264758 | |
| Translocation | Chr12 | 13244419 | Chr6 | 18212923 | |
| Translocation | Chr11 | 15116921 | Chr4 | 16850716 | 2 |
| Translocation | Chr11 | 15119400 | Chr4 | 16850755 | 2 |
| Translocation | Chr11 | 17538825 | Chr2 | 21868990 | 2 |
| Translocation | Chr11 | 17937586 | Chr6 | 7278777 | 2 |
| Translocation | Chr11 | 18052435 | Chr2 | 21869033 | 2 |
| Translocation | Chr11 | 18208023 | Chr6 | 25663354 |
