Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2085-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2085-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 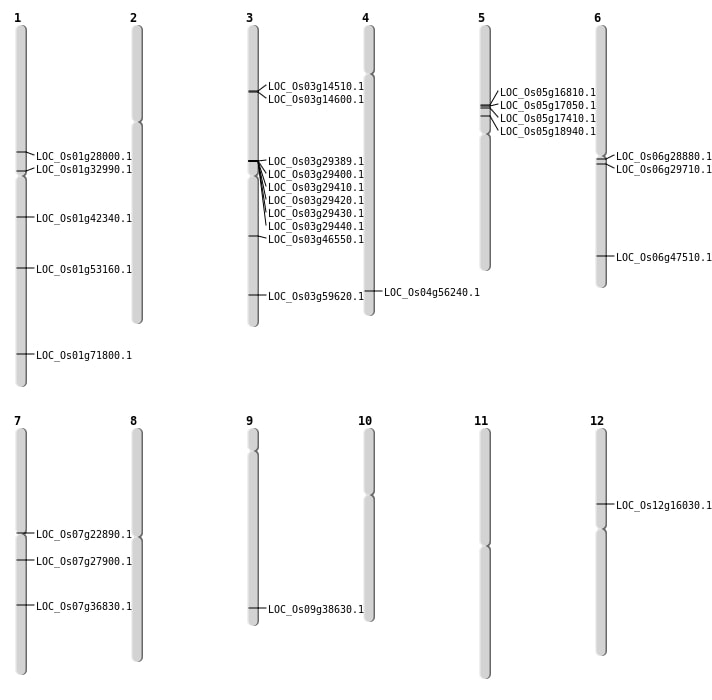
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14393417 | G-A | HET | ||
| SBS | Chr1 | 15660559 | T-C | HET | SPLICE_SITE_DONOR | LOC_Os01g28000 |
| SBS | Chr1 | 18119707 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g32990 |
| SBS | Chr1 | 24185770 | G-A | Homo | ||
| SBS | Chr1 | 30673891 | C-A | HET | ||
| SBS | Chr1 | 35841081 | T-C | Homo | ||
| SBS | Chr10 | 11334327 | G-A | Homo | ||
| SBS | Chr10 | 11495897 | T-C | HET | ||
| SBS | Chr10 | 12428448 | T-C | HET | ||
| SBS | Chr10 | 16412504 | C-A | HET | ||
| SBS | Chr10 | 22766738 | C-G | Homo | ||
| SBS | Chr10 | 2707179 | A-C | HET | ||
| SBS | Chr10 | 2957920 | A-G | HET | ||
| SBS | Chr10 | 4934277 | C-A | Homo | ||
| SBS | Chr11 | 10592020 | C-A | HET | ||
| SBS | Chr2 | 25612702 | A-T | HET | ||
| SBS | Chr2 | 27424698 | C-G | Homo | ||
| SBS | Chr2 | 3841407 | A-T | HET | ||
| SBS | Chr2 | 4632964 | A-T | HET | ||
| SBS | Chr2 | 6764463 | A-T | HET | ||
| SBS | Chr2 | 8289236 | A-C | HET | ||
| SBS | Chr3 | 15822283 | C-T | Homo | ||
| SBS | Chr3 | 1965196 | A-G | HET | ||
| SBS | Chr3 | 26340305 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46550 |
| SBS | Chr3 | 26645708 | T-C | HET | ||
| SBS | Chr3 | 7490459 | T-A | Homo | ||
| SBS | Chr4 | 12004120 | A-T | HET | ||
| SBS | Chr4 | 26769522 | T-C | HET | ||
| SBS | Chr4 | 2719133 | G-A | HET | ||
| SBS | Chr5 | 10991111 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18940 |
| SBS | Chr5 | 15602732 | G-T | HET | ||
| SBS | Chr5 | 15602733 | T-G | HET | ||
| SBS | Chr5 | 15602735 | G-T | HET | ||
| SBS | Chr5 | 18279445 | C-G | Homo | ||
| SBS | Chr5 | 28513326 | A-G | HET | ||
| SBS | Chr5 | 9737278 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g17050 |
| SBS | Chr5 | 9997702 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17410 |
| SBS | Chr6 | 16638126 | C-A | Homo | ||
| SBS | Chr6 | 23683360 | G-A | Homo | ||
| SBS | Chr6 | 25850936 | A-G | HET | ||
| SBS | Chr7 | 14301185 | C-A | HET | ||
| SBS | Chr7 | 16280609 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27900 |
| SBS | Chr7 | 28114885 | C-A | Homo | ||
| SBS | Chr8 | 24940376 | A-G | HET | ||
| SBS | Chr8 | 9196504 | G-T | HET | ||
| SBS | Chr8 | 9196505 | A-G | HET | ||
| SBS | Chr9 | 14324879 | G-C | Homo | ||
| SBS | Chr9 | 18271155 | A-G | HET | ||
| SBS | Chr9 | 22221759 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38630 |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2330256 | 2330259 | 3 | |
| Deletion | Chr8 | 6201309 | 6201490 | 181 | |
| Deletion | Chr3 | 7877320 | 7877333 | 13 | LOC_Os03g14510 |
| Deletion | Chr5 | 9578077 | 9578084 | 7 | LOC_Os05g16810 |
| Deletion | Chr3 | 13356192 | 13356194 | 2 | |
| Deletion | Chr3 | 16699500 | 16699519 | 19 | |
| Deletion | Chr3 | 16744001 | 16780000 | 35999 | 5 |
| Deletion | Chr11 | 24243978 | 24243991 | 13 | |
| Deletion | Chr7 | 24780101 | 24780103 | 2 | |
| Deletion | Chr2 | 26239410 | 26239419 | 9 | |
| Deletion | Chr2 | 27417641 | 27417643 | 2 | |
| Deletion | Chr1 | 30542967 | 30542968 | 1 | LOC_Os01g53160 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 21379748 | 21379749 | 2 | |
| Insertion | Chr3 | 7927151 | 7927183 | 33 | LOC_Os03g14600 |
| Insertion | Chr6 | 14476374 | 14476374 | 1 | |
| Insertion | Chr6 | 28763937 | 28763939 | 3 | LOC_Os06g47510 |
| Insertion | Chr8 | 20286334 | 20286367 | 34 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 1499149 | 1499252 | |
| Inversion | Chr7 | 12903849 | 12904063 | LOC_Os07g22890 |
| Inversion | Chr6 | 16477606 | 17061966 | 2 |
| Inversion | Chr1 | 23881643 | 24038464 | 2 |
| Inversion | Chr1 | 23881651 | 24038477 | 2 |
| Inversion | Chr11 | 24242427 | 24250207 | |
| Inversion | Chr3 | 33943841 | 33944041 | LOC_Os03g59620 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8228135 | Chr5 | 8546812 | |
| Translocation | Chr12 | 9135217 | Chr8 | 9992196 | LOC_Os12g16030 |
| Translocation | Chr7 | 10222779 | Chr1 | 1378166 | |
| Translocation | Chr6 | 16477602 | Chr3 | 16739600 | 2 |
| Translocation | Chr6 | 17061951 | Chr3 | 16744180 | LOC_Os06g29710 |
| Translocation | Chr7 | 22069268 | Chr6 | 25276925 | LOC_Os07g36830 |
| Translocation | Chr4 | 33532312 | Chr1 | 41587063 | 2 |
