Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2091-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2091-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 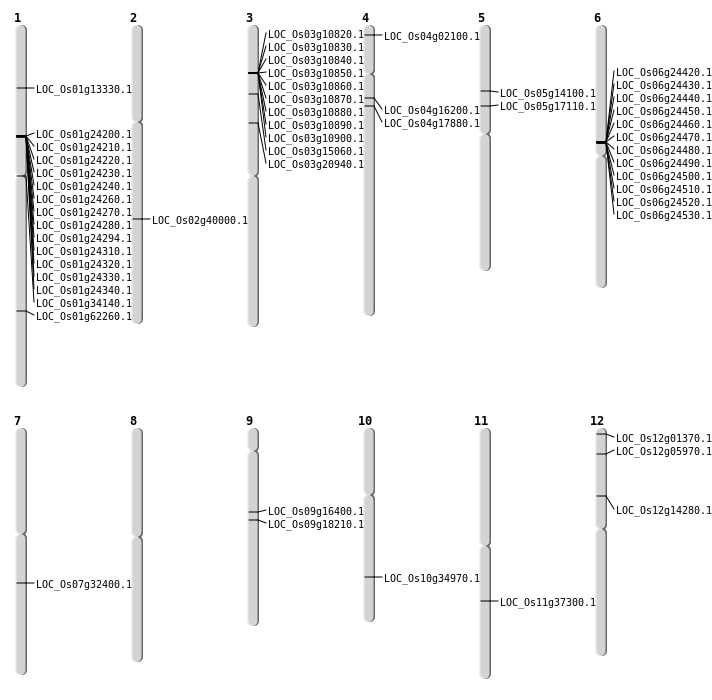
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16532109 | C-T | HET | ||
| SBS | Chr1 | 18810718 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34140 |
| SBS | Chr1 | 34457652 | C-A | HET | ||
| SBS | Chr10 | 12511563 | G-A | HET | ||
| SBS | Chr10 | 18939685 | A-G | HET | ||
| SBS | Chr10 | 22118239 | G-T | HET | ||
| SBS | Chr11 | 12306429 | G-A | HET | ||
| SBS | Chr11 | 13259231 | C-T | HET | ||
| SBS | Chr11 | 2090465 | G-A | HET | ||
| SBS | Chr11 | 21295364 | T-A | HET | ||
| SBS | Chr12 | 19238409 | T-C | HET | ||
| SBS | Chr12 | 20004164 | C-T | Homo | ||
| SBS | Chr12 | 8134000 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g14280 |
| SBS | Chr2 | 11189631 | G-A | HET | ||
| SBS | Chr2 | 11903725 | T-A | HET | ||
| SBS | Chr2 | 30584293 | C-G | HET | ||
| SBS | Chr2 | 3198292 | G-T | HET | ||
| SBS | Chr3 | 15099624 | C-T | HET | ||
| SBS | Chr3 | 31844232 | C-A | HET | ||
| SBS | Chr3 | 33682342 | T-C | Homo | ||
| SBS | Chr3 | 3391938 | T-C | HET | ||
| SBS | Chr3 | 36232485 | G-C | Homo | ||
| SBS | Chr4 | 18075771 | G-C | HET | ||
| SBS | Chr5 | 11728280 | G-A | HET | ||
| SBS | Chr5 | 25863315 | G-T | HET | ||
| SBS | Chr5 | 4477246 | G-A | HET | ||
| SBS | Chr5 | 5652015 | T-A | Homo | ||
| SBS | Chr5 | 5652016 | T-A | Homo | ||
| SBS | Chr5 | 5652060 | G-T | Homo | ||
| SBS | Chr5 | 6341043 | C-T | Homo | ||
| SBS | Chr5 | 7875984 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14100 |
| SBS | Chr6 | 14303570 | G-A | Homo | ||
| SBS | Chr6 | 15404010 | T-G | Homo | ||
| SBS | Chr6 | 24713079 | C-T | HET | ||
| SBS | Chr6 | 8620211 | T-A | HET | ||
| SBS | Chr8 | 16250490 | A-C | HET | ||
| SBS | Chr8 | 22603829 | C-T | HET | ||
| SBS | Chr8 | 27740012 | T-C | HET | ||
| SBS | Chr9 | 10031258 | G-A | HET | STOP_GAINED | LOC_Os09g16400 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2749766 | 2749767 | 1 | LOC_Os12g05970 |
| Deletion | Chr11 | 3186018 | 3186023 | 5 | |
| Deletion | Chr3 | 5544001 | 5600000 | 55999 | 9 |
| Deletion | Chr12 | 6859009 | 6859010 | 1 | |
| Deletion | Chr4 | 8796450 | 8796451 | 1 | LOC_Os04g16200 |
| Deletion | Chr8 | 9105698 | 9105704 | 6 | |
| Deletion | Chr12 | 9309081 | 9309082 | 1 | |
| Deletion | Chr5 | 9775635 | 9775638 | 3 | LOC_Os05g17110 |
| Deletion | Chr4 | 9824713 | 9824714 | 1 | LOC_Os04g17880 |
| Deletion | Chr9 | 11161921 | 11161944 | 23 | LOC_Os09g18210 |
| Deletion | Chr6 | 12657438 | 12657444 | 6 | |
| Deletion | Chr1 | 13635001 | 13690000 | 54999 | 9 |
| Deletion | Chr1 | 13691001 | 13717000 | 25999 | 5 |
| Deletion | Chr6 | 14305001 | 14385000 | 79999 | 12 |
| Deletion | Chr11 | 14602721 | 14602730 | 9 | |
| Deletion | Chr10 | 14719507 | 14719513 | 6 | |
| Deletion | Chr5 | 18038658 | 18038662 | 4 | |
| Deletion | Chr11 | 21024699 | 21024701 | 2 | |
| Deletion | Chr3 | 36235333 | 36235335 | 2 | |
| Deletion | Chr1 | 37488954 | 37488956 | 2 | |
| Deletion | Chr1 | 38649300 | 38649301 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 18187624 | 18187624 | 1 | |
| Insertion | Chr5 | 29472664 | 29472665 | 2 | |
| Insertion | Chr9 | 16146205 | 16146218 | 14 |
Inversions: 13
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7432461 | 7432591 | LOC_Os01g13330 |
| Inversion | Chr8 | 8981228 | 9474207 | |
| Inversion | Chr8 | 8981232 | 9474732 | |
| Inversion | Chr8 | 13390230 | 13390355 | |
| Inversion | Chr6 | 13824594 | 14385178 | |
| Inversion | Chr6 | 14400258 | 14400398 | |
| Inversion | Chr10 | 18662092 | 18662193 | LOC_Os10g34970 |
| Inversion | Chr7 | 19265943 | 19266404 | LOC_Os07g32400 |
| Inversion | Chr4 | 21069241 | 21069458 | |
| Inversion | Chr11 | 21412876 | 22027876 | 2 |
| Inversion | Chr11 | 21412877 | 22027876 | 2 |
| Inversion | Chr3 | 25288818 | 25289097 | |
| Inversion | Chr1 | 31523263 | 31523454 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 229497 | Chr11 | 204242 | LOC_Os12g01370 |
| Translocation | Chr11 | 1111606 | Chr1 | 24516348 | |
| Translocation | Chr11 | 1111607 | Chr1 | 24516339 | |
| Translocation | Chr10 | 2469440 | Chr4 | 679680 | 2 |
| Translocation | Chr6 | 3033845 | Chr3 | 8228323 | 2 |
| Translocation | Chr3 | 11878381 | Chr1 | 37612656 | LOC_Os03g20940 |
| Translocation | Chr3 | 11881884 | Chr1 | 37612796 | |
| Translocation | Chr6 | 14305127 | Chr3 | 11881867 | LOC_Os06g24420 |
| Translocation | Chr3 | 14720376 | Chr2 | 24201916 | |
| Translocation | Chr3 | 14720377 | Chr2 | 24200634 | 2 |
| Translocation | Chr9 | 17421077 | Chr1 | 36029808 | 2 |
| Translocation | Chr6 | 24088199 | Chr2 | 25837297 |
