Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2129-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2129-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 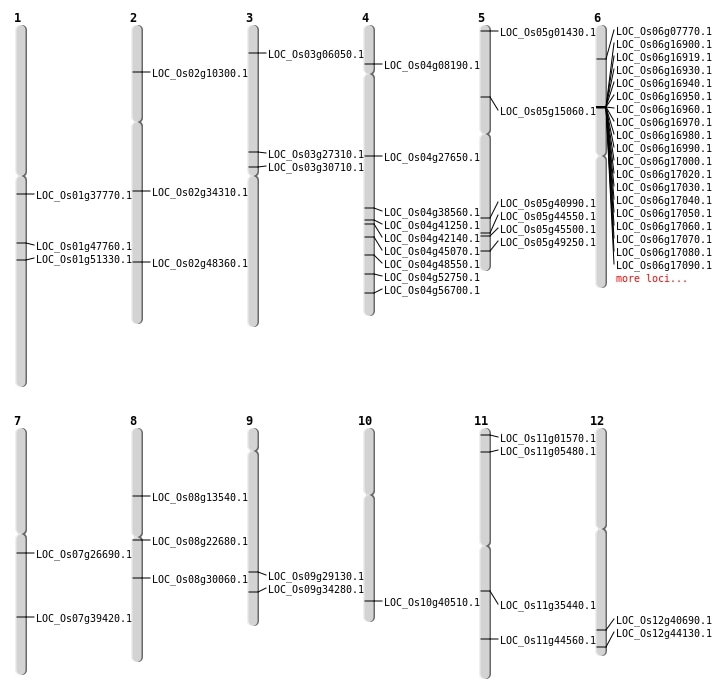
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21003504 | A-G | Homo | ||
| SBS | Chr1 | 21300109 | A-T | HET | ||
| SBS | Chr1 | 8564741 | A-G | HET | ||
| SBS | Chr10 | 22066081 | G-A | HET | ||
| SBS | Chr10 | 547227 | G-A | Homo | ||
| SBS | Chr11 | 1737721 | A-G | HET | ||
| SBS | Chr11 | 4095205 | C-T | HET | ||
| SBS | Chr11 | 5453446 | G-A | HET | ||
| SBS | Chr11 | 5740953 | G-A | HET | ||
| SBS | Chr11 | 7552677 | G-T | Homo | ||
| SBS | Chr12 | 17095009 | C-T | HET | ||
| SBS | Chr12 | 20982105 | C-T | Homo | ||
| SBS | Chr12 | 25191985 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g40690 |
| SBS | Chr12 | 25191986 | C-T | Homo | ||
| SBS | Chr12 | 6728569 | C-T | HET | ||
| SBS | Chr2 | 118722 | T-C | HET | ||
| SBS | Chr2 | 15335012 | G-A | HET | ||
| SBS | Chr2 | 31101301 | G-A | HET | ||
| SBS | Chr2 | 31101302 | T-G | HET | ||
| SBS | Chr2 | 5830449 | T-C | HET | ||
| SBS | Chr4 | 10840220 | T-C | HET | ||
| SBS | Chr4 | 16341999 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g27650 |
| SBS | Chr4 | 19149670 | C-T | HET | ||
| SBS | Chr4 | 24947576 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g42140 |
| SBS | Chr4 | 25290349 | C-T | HET | ||
| SBS | Chr5 | 8654875 | C-T | Homo | ||
| SBS | Chr6 | 22124585 | C-T | Homo | SPLICE_SITE_ACCEPTOR | LOC_Os06g37420 |
| SBS | Chr6 | 23022546 | T-A | Homo | ||
| SBS | Chr6 | 23292428 | A-G | Homo | ||
| SBS | Chr6 | 26581751 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44070 |
| SBS | Chr6 | 8763018 | C-A | HET | ||
| SBS | Chr7 | 925081 | T-C | Homo | ||
| SBS | Chr8 | 12282589 | T-C | HET | ||
| SBS | Chr8 | 17259784 | A-T | HET | ||
| SBS | Chr8 | 2466567 | C-G | HET | ||
| SBS | Chr8 | 26954977 | G-A | HET | ||
| SBS | Chr8 | 3604014 | T-C | HET | ||
| SBS | Chr8 | 8049933 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g13540 |
| SBS | Chr9 | 11041907 | G-T | HET | ||
| SBS | Chr9 | 14356202 | A-G | HET | ||
| SBS | Chr9 | 8099038 | C-A | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 329742 | 329748 | 6 | LOC_Os11g01570 |
| Deletion | Chr4 | 2766327 | 2766329 | 2 | |
| Deletion | Chr8 | 4727063 | 4727064 | 1 | |
| Deletion | Chr7 | 9443682 | 9443683 | 1 | |
| Deletion | Chr6 | 9795001 | 9956000 | 160999 | 26 |
| Deletion | Chr2 | 10756220 | 10756221 | 1 | |
| Deletion | Chr4 | 12986778 | 12986781 | 3 | |
| Deletion | Chr3 | 13467920 | 13467921 | 1 | |
| Deletion | Chr4 | 17450648 | 17450649 | 1 | |
| Deletion | Chr12 | 17458537 | 17458540 | 3 | |
| Deletion | Chr3 | 17500003 | 17500004 | 1 | LOC_Os03g30710 |
| Deletion | Chr5 | 21755976 | 21755978 | 2 | |
| Deletion | Chr3 | 22841859 | 22841867 | 8 | |
| Deletion | Chr1 | 23122096 | 23122108 | 12 | |
| Deletion | Chr8 | 23789810 | 23789827 | 17 | |
| Deletion | Chr8 | 26717558 | 26717559 | 1 | |
| Deletion | Chr11 | 27141710 | 27141724 | 14 | |
| Deletion | Chr4 | 28412089 | 28412091 | 2 | |
| Deletion | Chr4 | 33812130 | 33812131 | 1 | LOC_Os04g56700 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23047852 | 23047853 | 2 | |
| Insertion | Chr1 | 31479258 | 31479259 | 2 | |
| Insertion | Chr10 | 12765858 | 12765859 | 2 | |
| Insertion | Chr4 | 1274218 | 1274220 | 3 | |
| Insertion | Chr8 | 13642396 | 13642396 | 1 | LOC_Os08g22680 |
| Insertion | Chr8 | 8733971 | 8733971 | 1 |
Inversions: 57
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 249197 | 249460 | LOC_Os05g01430 |
| Inversion | Chr9 | 2096813 | 2097180 | |
| Inversion | Chr11 | 2466816 | 2467085 | LOC_Os11g05480 |
| Inversion | Chr8 | 2543550 | 2543718 | |
| Inversion | Chr3 | 3026746 | 3027095 | LOC_Os03g06050 |
| Inversion | Chr6 | 3766375 | 3766624 | LOC_Os06g07770 |
| Inversion | Chr9 | 4177046 | 4177291 | |
| Inversion | Chr4 | 4353175 | 4353500 | LOC_Os04g08190 |
| Inversion | Chr2 | 5412191 | 5412325 | LOC_Os02g10300 |
| Inversion | Chr2 | 5894166 | 5894498 | |
| Inversion | Chr7 | 6083727 | 6084089 | |
| Inversion | Chr5 | 8551261 | 8551364 | 2 |
| Inversion | Chr1 | 10400354 | 10400786 | |
| Inversion | Chr7 | 11547762 | 11548022 | |
| Inversion | Chr4 | 13243008 | 13243198 | |
| Inversion | Chr4 | 14069658 | 14069779 | |
| Inversion | Chr7 | 15406419 | 15406583 | 2 |
| Inversion | Chr3 | 15436025 | 15436240 | |
| Inversion | Chr3 | 15647690 | 15647848 | LOC_Os03g27310 |
| Inversion | Chr12 | 15994306 | 15994774 | |
| Inversion | Chr9 | 17704917 | 17705144 | LOC_Os09g29130 |
| Inversion | Chr8 | 18480091 | 18480303 | LOC_Os08g30060 |
| Inversion | Chr5 | 19171982 | 19172114 | |
| Inversion | Chr10 | 19559818 | 19559994 | |
| Inversion | Chr9 | 20236561 | 20236663 | LOC_Os09g34280 |
| Inversion | Chr2 | 20530011 | 20530130 | LOC_Os02g34310 |
| Inversion | Chr11 | 20765444 | 20765873 | LOC_Os11g35440 |
| Inversion | Chr1 | 21127931 | 21128244 | LOC_Os01g37770 |
| Inversion | Chr10 | 21690272 | 21690718 | 2 |
| Inversion | Chr8 | 21832090 | 21832245 | |
| Inversion | Chr10 | 21872168 | 21872280 | |
| Inversion | Chr4 | 22910244 | 22910411 | 2 |
| Inversion | Chr5 | 23461523 | 23461797 | |
| Inversion | Chr7 | 23612713 | 23612924 | 2 |
| Inversion | Chr5 | 24028787 | 24029014 | LOC_Os05g40990 |
| Inversion | Chr4 | 24472422 | 24472522 | LOC_Os04g41250 |
| Inversion | Chr1 | 25092253 | 25092533 | |
| Inversion | Chr5 | 25915536 | 25915987 | LOC_Os05g44550 |
| Inversion | Chr5 | 26384546 | 26384816 | 2 |
| Inversion | Chr4 | 26667804 | 26667996 | LOC_Os04g45070 |
| Inversion | Chr11 | 26945585 | 26945889 | LOC_Os11g44560 |
| Inversion | Chr1 | 27326379 | 27326568 | LOC_Os01g47760 |
| Inversion | Chr12 | 27377051 | 27377189 | LOC_Os12g44130 |
| Inversion | Chr11 | 28129772 | 28130316 | |
| Inversion | Chr5 | 28253745 | 28254166 | LOC_Os05g49250 |
| Inversion | Chr4 | 28816553 | 28816940 | |
| Inversion | Chr4 | 28940446 | 28940605 | LOC_Os04g48550 |
| Inversion | Chr6 | 28951040 | 28951481 | LOC_Os06g47834 |
| Inversion | Chr4 | 28979057 | 28979199 | |
| Inversion | Chr1 | 29522811 | 29523055 | LOC_Os01g51330 |
| Inversion | Chr2 | 29606242 | 29606645 | LOC_Os02g48360 |
| Inversion | Chr3 | 30050008 | 30050244 | |
| Inversion | Chr4 | 31410507 | 31410899 | LOC_Os04g52750 |
| Inversion | Chr3 | 34183301 | 34183709 | |
| Inversion | Chr2 | 34346430 | 34346714 | |
| Inversion | Chr3 | 35786861 | 35787219 | |
| Inversion | Chr1 | 40678122 | 40678460 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 26002703 | Chr6 | 9594568 |
