Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2152-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2152-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 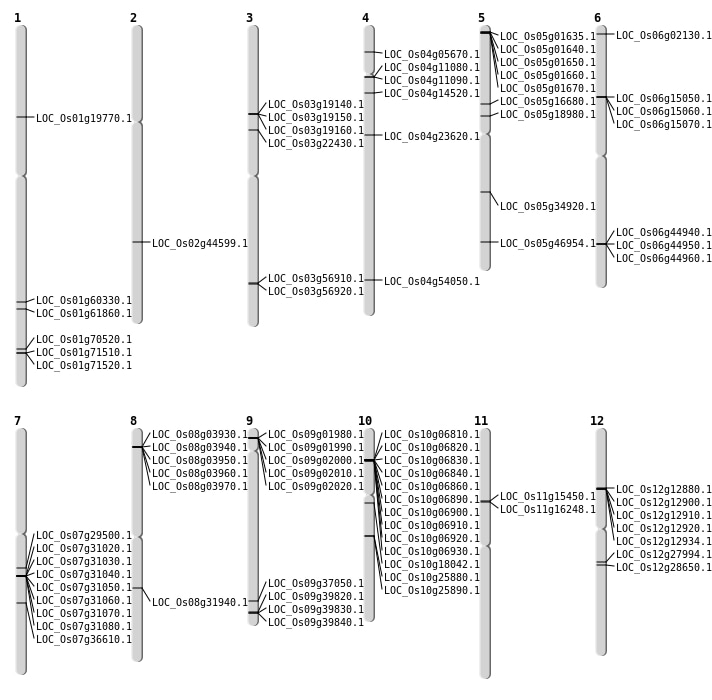
|
| Variant Information |
|---|
Single base substitutions: 20
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11222885 | T-G | HET | SPLICE_SITE_ACCEPTOR | LOC_Os01g19770 |
| SBS | Chr1 | 29835432 | C-A | Homo | ||
| SBS | Chr1 | 34900164 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60330 |
| SBS | Chr11 | 15979548 | G-A | HET | ||
| SBS | Chr11 | 3282349 | T-C | HET | ||
| SBS | Chr3 | 12433111 | C-T | HET | ||
| SBS | Chr3 | 21449338 | G-A | HET | ||
| SBS | Chr4 | 24712352 | C-T | HET | ||
| SBS | Chr4 | 2898224 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05670 |
| SBS | Chr5 | 11016069 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18980 |
| SBS | Chr5 | 27171128 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g46954 |
| SBS | Chr6 | 24069430 | A-C | Homo | ||
| SBS | Chr6 | 26660957 | A-G | HET | ||
| SBS | Chr6 | 634635 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g02130 |
| SBS | Chr6 | 7179348 | C-A | Homo | ||
| SBS | Chr7 | 14289804 | G-A | HET | ||
| SBS | Chr7 | 21905176 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36610 |
| SBS | Chr7 | 6663831 | C-T | HET | ||
| SBS | Chr8 | 12519489 | T-G | Homo | ||
| SBS | Chr8 | 782223 | A-G | Homo |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 378001 | 400000 | 21999 | 5 |
| Deletion | Chr9 | 685001 | 706000 | 20999 | 5 |
| Deletion | Chr8 | 781993 | 782010 | 17 | |
| Deletion | Chr8 | 1873001 | 1897000 | 23999 | 5 |
| Deletion | Chr10 | 3545001 | 3617000 | 71999 | 10 |
| Deletion | Chr4 | 6031001 | 6041000 | 9999 | 2 |
| Deletion | Chr6 | 7102472 | 7102479 | 7 | |
| Deletion | Chr12 | 7124001 | 7168000 | 43999 | 5 |
| Deletion | Chr6 | 8530001 | 8551000 | 20999 | 3 |
| Deletion | Chr1 | 9032805 | 9032806 | 1 | |
| Deletion | Chr1 | 10631644 | 10631645 | 1 | |
| Deletion | Chr3 | 10719001 | 10743000 | 23999 | 3 |
| Deletion | Chr12 | 11509869 | 11509870 | 1 | |
| Deletion | Chr11 | 12069463 | 12069464 | 1 | |
| Deletion | Chr8 | 13029536 | 13029543 | 7 | |
| Deletion | Chr10 | 13405001 | 13421000 | 15999 | 2 |
| Deletion | Chr12 | 16938190 | 16938192 | 2 | LOC_Os12g28650 |
| Deletion | Chr7 | 18369001 | 18405000 | 35999 | 7 |
| Deletion | Chr5 | 20750746 | 20750750 | 4 | LOC_Os05g34920 |
| Deletion | Chr5 | 21256368 | 21256369 | 1 | |
| Deletion | Chr9 | 22825001 | 22844000 | 18999 | 3 |
| Deletion | Chr12 | 25692196 | 25692200 | 4 | |
| Deletion | Chr6 | 27175001 | 27198000 | 22999 | 3 |
| Deletion | Chr3 | 28608197 | 28608199 | 2 | |
| Deletion | Chr3 | 32411001 | 32422000 | 10999 | 2 |
| Deletion | Chr1 | 35792176 | 35792179 | 3 | LOC_Os01g61860 |
| Deletion | Chr1 | 41407001 | 41424000 | 16999 | 2 |
Insertions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23145025 | 23145025 | 1 | |
| Insertion | Chr1 | 27809054 | 27809054 | 1 | |
| Insertion | Chr10 | 9144888 | 9144888 | 1 | LOC_Os10g18042 |
| Insertion | Chr11 | 13493795 | 13493795 | 1 | |
| Insertion | Chr12 | 9229235 | 9229235 | 1 | |
| Insertion | Chr3 | 10745204 | 10745204 | 1 | |
| Insertion | Chr3 | 14126142 | 14126142 | 1 | |
| Insertion | Chr3 | 3222577 | 3222577 | 1 | |
| Insertion | Chr4 | 15320793 | 15320793 | 1 | |
| Insertion | Chr4 | 5447379 | 5447379 | 1 | |
| Insertion | Chr4 | 8143986 | 8144006 | 21 | |
| Insertion | Chr5 | 18369178 | 18369179 | 2 | |
| Insertion | Chr5 | 9483217 | 9483234 | 18 | LOC_Os05g16680 |
| Insertion | Chr7 | 12486198 | 12486199 | 2 | |
| Insertion | Chr7 | 15467387 | 15467387 | 1 | |
| Insertion | Chr7 | 17320556 | 17320556 | 1 | LOC_Os07g29500 |
| Insertion | Chr8 | 23298558 | 23298588 | 31 | |
| Insertion | Chr8 | 24362510 | 24362510 | 1 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 8758810 | 8938868 | 2 |
| Inversion | Chr9 | 9739410 | 9739675 | |
| Inversion | Chr12 | 16504747 | 16505102 | LOC_Os12g27994 |
| Inversion | Chr2 | 19073093 | 19073394 | |
| Inversion | Chr9 | 21371763 | 21372153 | LOC_Os09g37050 |
| Inversion | Chr1 | 25249683 | 25249953 | |
| Inversion | Chr3 | 31521165 | 31521417 | |
| Inversion | Chr4 | 32084501 | 32209635 | 2 |
| Inversion | Chr2 | 32700585 | 32700704 | LOC_Os02g53450 |
Translocations: 18
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 4244184 | Chr2 | 6526410 | |
| Translocation | Chr5 | 4954614 | Chr4 | 857584 | |
| Translocation | Chr12 | 6215582 | Chr2 | 1697509 | |
| Translocation | Chr12 | 9802786 | Chr10 | 20738719 | |
| Translocation | Chr3 | 12858565 | Chr2 | 27027477 | 2 |
| Translocation | Chr10 | 12950318 | Chr8 | 14202216 | |
| Translocation | Chr9 | 13150340 | Chr4 | 13523968 | 2 |
| Translocation | Chr9 | 13150371 | Chr4 | 13439748 | |
| Translocation | Chr10 | 14180847 | Chr1 | 26307386 | |
| Translocation | Chr11 | 14589444 | Chr4 | 8131957 | 2 |
| Translocation | Chr3 | 17136806 | Chr2 | 24543571 | |
| Translocation | Chr5 | 18271869 | Chr4 | 28907055 | |
| Translocation | Chr11 | 19220031 | Chr9 | 8149217 | |
| Translocation | Chr8 | 19804265 | Chr7 | 1870478 | LOC_Os08g31940 |
| Translocation | Chr5 | 20654989 | Chr1 | 40847961 | 2 |
| Translocation | Chr5 | 20669340 | Chr1 | 40847958 | 2 |
| Translocation | Chr11 | 22131407 | Chr7 | 11905199 | |
| Translocation | Chr11 | 27827160 | Chr6 | 25898014 |
