Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2157-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2157-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 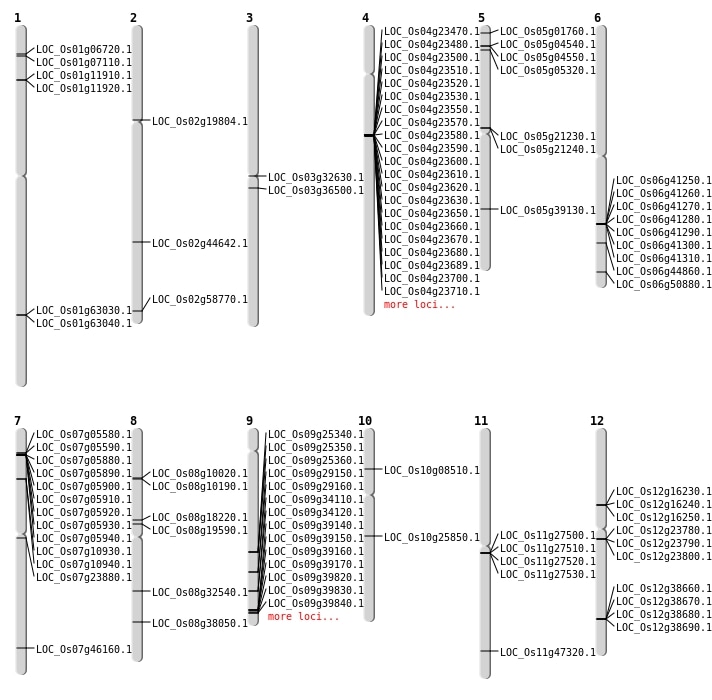
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13850172 | C-A | Homo | ||
| SBS | Chr10 | 1912240 | G-A | HET | ||
| SBS | Chr11 | 18836550 | C-T | Homo | ||
| SBS | Chr11 | 20386667 | C-A | Homo | ||
| SBS | Chr11 | 28240714 | G-A | Homo | ||
| SBS | Chr12 | 12068187 | G-A | HET | ||
| SBS | Chr12 | 19421957 | C-T | Homo | ||
| SBS | Chr12 | 1965598 | A-G | HET | ||
| SBS | Chr12 | 9716659 | T-A | HET | ||
| SBS | Chr2 | 19171304 | A-G | HET | ||
| SBS | Chr2 | 32622731 | G-A | HET | ||
| SBS | Chr3 | 6314468 | C-T | HET | ||
| SBS | Chr3 | 6355800 | G-A | HET | ||
| SBS | Chr4 | 20421661 | A-G | Homo | ||
| SBS | Chr5 | 184081 | G-A | Homo | ||
| SBS | Chr5 | 22949094 | A-T | Homo | STOP_GAINED | LOC_Os05g39130 |
| SBS | Chr6 | 12259604 | C-G | HET | ||
| SBS | Chr6 | 12259608 | C-T | HET | ||
| SBS | Chr6 | 6851803 | C-G | HET | ||
| SBS | Chr7 | 13522317 | T-A | HET | ||
| SBS | Chr7 | 22885158 | T-G | HET | ||
| SBS | Chr7 | 27021735 | A-G | HET | ||
| SBS | Chr8 | 10882041 | T-C | HET | ||
| SBS | Chr8 | 16890726 | T-A | HET | ||
| SBS | Chr8 | 24615413 | G-T | HET | ||
| SBS | Chr8 | 24615414 | A-T | HET | ||
| SBS | Chr8 | 5819444 | A-T | HET | ||
| SBS | Chr9 | 20822657 | C-T | HET |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 611133 | 611145 | 12 | |
| Deletion | Chr5 | 2105001 | 2124000 | 18999 | 2 |
| Deletion | Chr7 | 2615001 | 2628000 | 12999 | 2 |
| Deletion | Chr7 | 2825001 | 2882000 | 56999 | 7 |
| Deletion | Chr8 | 5795001 | 5808000 | 12999 | LOC_Os08g10020 |
| Deletion | Chr8 | 5910928 | 5910938 | 10 | LOC_Os08g10190 |
| Deletion | Chr7 | 5983001 | 5996000 | 12999 | 2 |
| Deletion | Chr1 | 6463001 | 6473000 | 9999 | 2 |
| Deletion | Chr6 | 6650207 | 6650217 | 10 | |
| Deletion | Chr9 | 8516720 | 8516723 | 3 | |
| Deletion | Chr12 | 9278001 | 9292000 | 13999 | 4 |
| Deletion | Chr1 | 9459968 | 9459984 | 16 | |
| Deletion | Chr1 | 10270725 | 10270727 | 2 | |
| Deletion | Chr11 | 10572591 | 10572592 | 1 | |
| Deletion | Chr8 | 11166774 | 11166775 | 1 | LOC_Os08g18220 |
| Deletion | Chr5 | 12557001 | 12580000 | 22999 | 2 |
| Deletion | Chr4 | 13417001 | 13668000 | 250999 | 32 |
| Deletion | Chr12 | 13509001 | 13528000 | 18999 | 3 |
| Deletion | Chr11 | 13675632 | 13675633 | 1 | |
| Deletion | Chr11 | 14600982 | 14600983 | 1 | |
| Deletion | Chr5 | 14904708 | 14904709 | 1 | |
| Deletion | Chr9 | 15172001 | 15201000 | 28999 | 3 |
| Deletion | Chr12 | 15239088 | 15239089 | 1 | |
| Deletion | Chr11 | 15818001 | 15840000 | 21999 | 4 |
| Deletion | Chr9 | 17708001 | 17732000 | 23999 | 2 |
| Deletion | Chr4 | 18346001 | 18361000 | 14999 | 2 |
| Deletion | Chr9 | 20134001 | 20146000 | 11999 | 2 |
| Deletion | Chr9 | 22466001 | 22499000 | 32999 | 4 |
| Deletion | Chr9 | 22823001 | 22852000 | 28999 | 4 |
| Deletion | Chr12 | 23741001 | 23765000 | 23999 | 4 |
| Deletion | Chr6 | 24689001 | 24717000 | 27999 | 7 |
| Deletion | Chr8 | 26854997 | 26855000 | 3 | |
| Deletion | Chr6 | 27100001 | 27116000 | 15999 | LOC_Os06g44860 |
| Deletion | Chr1 | 36505001 | 36526000 | 20999 | 2 |
Insertions: 5
Inversions: 35
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 455392 | 455740 | LOC_Os05g01760 |
| Inversion | Chr5 | 2623353 | 2623488 | LOC_Os05g05320 |
| Inversion | Chr2 | 3161019 | 3161381 | |
| Inversion | Chr1 | 3177992 | 3178388 | LOC_Os01g06720 |
| Inversion | Chr1 | 3352719 | 3353044 | LOC_Os01g07110 |
| Inversion | Chr1 | 4457184 | 4457404 | |
| Inversion | Chr2 | 6668299 | 6668652 | |
| Inversion | Chr9 | 8038696 | 8038822 | |
| Inversion | Chr12 | 9166738 | 9167218 | |
| Inversion | Chr5 | 11006968 | 11007541 | |
| Inversion | Chr2 | 11578163 | 11578539 | 2 |
| Inversion | Chr8 | 11708086 | 11708374 | LOC_Os08g19590 |
| Inversion | Chr9 | 12018937 | 12019167 | |
| Inversion | Chr1 | 12497700 | 12497885 | |
| Inversion | Chr10 | 13402131 | 13552387 | LOC_Os10g25850 |
| Inversion | Chr7 | 13491146 | 13491384 | LOC_Os07g23880 |
| Inversion | Chr5 | 17360964 | 17361343 | |
| Inversion | Chr7 | 18670041 | 18692960 | |
| Inversion | Chr7 | 18674261 | 18692961 | |
| Inversion | Chr3 | 18687312 | 18687711 | LOC_Os03g32630 |
| Inversion | Chr8 | 20158543 | 20158819 | LOC_Os08g32540 |
| Inversion | Chr3 | 20216759 | 20216920 | LOC_Os03g36500 |
| Inversion | Chr11 | 20548021 | 20548362 | |
| Inversion | Chr4 | 20652825 | 20653140 | |
| Inversion | Chr4 | 22348224 | 22348493 | LOC_Os04g37550 |
| Inversion | Chr10 | 22444845 | 22449576 | |
| Inversion | Chr4 | 23113166 | 23679155 | 2 |
| Inversion | Chr8 | 23697578 | 24109768 | 2 |
| Inversion | Chr11 | 26412323 | 26412646 | |
| Inversion | Chr2 | 27058309 | 27058706 | LOC_Os02g44642 |
| Inversion | Chr7 | 27549416 | 27549600 | LOC_Os07g46160 |
| Inversion | Chr3 | 27608362 | 27608549 | |
| Inversion | Chr3 | 31298039 | 31298574 | |
| Inversion | Chr4 | 31586459 | 31586728 | 2 |
| Inversion | Chr4 | 34440284 | 34440514 | LOC_Os04g57840 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2466352 | Chr6 | 30778799 | 2 |
| Translocation | Chr12 | 2466359 | Chr6 | 30778800 | 2 |
| Translocation | Chr7 | 4450793 | Chr6 | 30983134 | |
| Translocation | Chr10 | 4614010 | Chr6 | 10862726 | LOC_Os10g08510 |
| Translocation | Chr12 | 8680183 | Chr11 | 27722254 | |
| Translocation | Chr9 | 12526326 | Chr2 | 35903250 | 2 |
| Translocation | Chr9 | 12531575 | Chr2 | 35903231 | 2 |
| Translocation | Chr11 | 13220749 | Chr5 | 29713108 | |
| Translocation | Chr3 | 13646418 | Chr1 | 33549357 | |
| Translocation | Chr11 | 28429707 | Chr3 | 16633154 | |
| Translocation | Chr11 | 28467263 | Chr8 | 23697565 | LOC_Os11g47320 |
