Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2166-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2166-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 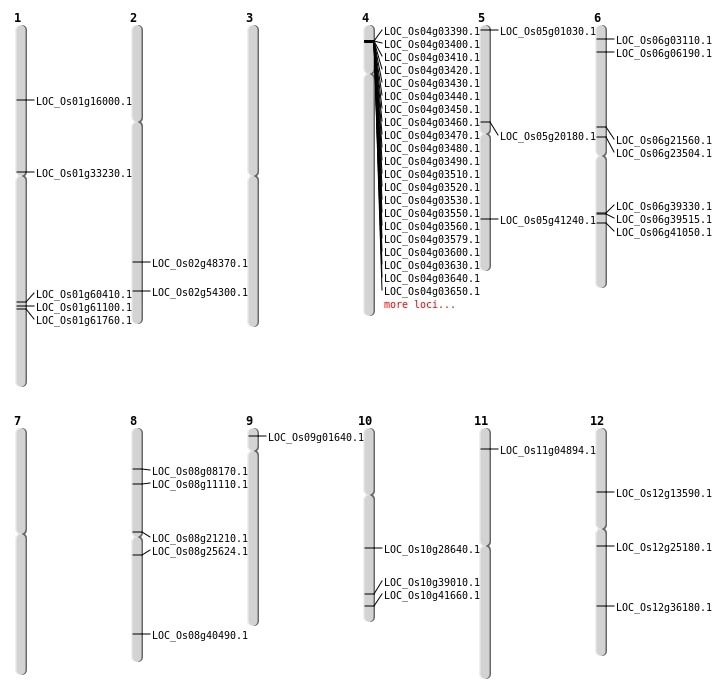
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23038029 | A-T | Homo | ||
| SBS | Chr1 | 24637362 | G-A | HET | ||
| SBS | Chr1 | 26224609 | A-G | HET | ||
| SBS | Chr1 | 30298239 | T-C | Homo | ||
| SBS | Chr1 | 38092381 | T-C | HET | ||
| SBS | Chr1 | 41937742 | T-C | HET | ||
| SBS | Chr1 | 7019838 | T-A | HET | ||
| SBS | Chr10 | 8944938 | T-C | HET | ||
| SBS | Chr11 | 19305168 | A-T | Homo | ||
| SBS | Chr11 | 2087410 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g04894 |
| SBS | Chr11 | 2087414 | G-C | HET | ||
| SBS | Chr11 | 25507958 | C-T | HET | ||
| SBS | Chr11 | 6469086 | T-A | Homo | ||
| SBS | Chr12 | 23335812 | C-T | HET | ||
| SBS | Chr12 | 6545635 | T-C | HET | ||
| SBS | Chr2 | 12537487 | C-T | Homo | ||
| SBS | Chr2 | 2175584 | G-A | HET | ||
| SBS | Chr2 | 33299792 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g54300 |
| SBS | Chr2 | 414216 | G-T | HET | ||
| SBS | Chr2 | 414217 | A-T | HET | ||
| SBS | Chr3 | 11854228 | G-A | HET | ||
| SBS | Chr3 | 1556797 | A-T | HET | ||
| SBS | Chr3 | 21370545 | G-A | HET | ||
| SBS | Chr3 | 24242482 | C-T | HET | ||
| SBS | Chr3 | 28816969 | T-C | HET | ||
| SBS | Chr3 | 29051881 | G-C | HET | ||
| SBS | Chr3 | 31165513 | T-A | HET | ||
| SBS | Chr3 | 8112683 | G-A | HET | ||
| SBS | Chr4 | 1277505 | T-C | HET | ||
| SBS | Chr4 | 14586968 | T-C | Homo | ||
| SBS | Chr4 | 25108756 | C-T | HET | ||
| SBS | Chr4 | 34248074 | C-T | HET | ||
| SBS | Chr4 | 35264914 | C-T | HET | ||
| SBS | Chr4 | 4649084 | A-T | HET | ||
| SBS | Chr5 | 11812573 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20180 |
| SBS | Chr5 | 14194641 | C-A | HET | ||
| SBS | Chr5 | 15271549 | T-A | HET | ||
| SBS | Chr5 | 17731024 | G-A | HET | ||
| SBS | Chr5 | 21520933 | T-C | HET | ||
| SBS | Chr5 | 9836368 | G-A | HET | ||
| SBS | Chr6 | 1146163 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g03110 |
| SBS | Chr6 | 1146164 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g03110 |
| SBS | Chr6 | 26569036 | A-G | Homo | ||
| SBS | Chr6 | 2866718 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g06190 |
| SBS | Chr7 | 12960621 | C-A | HET | ||
| SBS | Chr7 | 3753480 | G-A | HET | ||
| SBS | Chr8 | 16045578 | C-A | HET | ||
| SBS | Chr8 | 18839687 | G-A | HET | ||
| SBS | Chr8 | 21995868 | T-A | HET | ||
| SBS | Chr8 | 22247717 | A-T | HET | ||
| SBS | Chr8 | 22247719 | G-T | HET | ||
| SBS | Chr8 | 5506033 | C-G | HET |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1454001 | 1510000 | 55999 | 11 |
| Deletion | Chr4 | 1514001 | 1577000 | 62999 | 7 |
| Deletion | Chr4 | 1590001 | 1807000 | 216999 | 31 |
| Deletion | Chr9 | 11243501 | 11243502 | 1 | |
| Deletion | Chr9 | 11332386 | 11332388 | 2 | |
| Deletion | Chr6 | 12434849 | 12434853 | 4 | LOC_Os06g21560 |
| Deletion | Chr2 | 12531434 | 12531441 | 7 | |
| Deletion | Chr7 | 17108965 | 17109080 | 115 | |
| Deletion | Chr1 | 17165437 | 17165449 | 12 | |
| Deletion | Chr7 | 19004562 | 19004563 | 1 | |
| Deletion | Chr4 | 19724764 | 19724765 | 1 | LOC_Os04g32720 |
| Deletion | Chr2 | 21353744 | 21353745 | 1 | |
| Deletion | Chr1 | 23973902 | 23973903 | 1 | |
| Deletion | Chr6 | 24517360 | 24517362 | 2 | LOC_Os06g41050 |
| Deletion | Chr7 | 26024302 | 26024303 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14946944 | 14946946 | 3 | |
| Insertion | Chr2 | 25847793 | 25847794 | 2 | |
| Insertion | Chr5 | 5266970 | 5266971 | 2 | |
| Insertion | Chr6 | 22495131 | 22495149 | 19 |
Inversions: 38
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 62940 | 63208 | LOC_Os05g01030 |
| Inversion | Chr9 | 457233 | 457412 | LOC_Os09g01640 |
| Inversion | Chr4 | 1604550 | 1604840 | |
| Inversion | Chr5 | 2350173 | 2350507 | |
| Inversion | Chr6 | 2555191 | 2555330 | |
| Inversion | Chr4 | 3211404 | 3211680 | |
| Inversion | Chr9 | 5070002 | 5070148 | |
| Inversion | Chr8 | 6536435 | 6536561 | LOC_Os08g11110 |
| Inversion | Chr12 | 7640010 | 7640219 | LOC_Os12g13590 |
| Inversion | Chr1 | 9005130 | 9005303 | LOC_Os01g16000 |
| Inversion | Chr3 | 10656802 | 10656935 | |
| Inversion | Chr1 | 11278945 | 11279147 | |
| Inversion | Chr5 | 12288221 | 12288556 | |
| Inversion | Chr12 | 14452973 | 14453285 | 2 |
| Inversion | Chr10 | 14929288 | 14929440 | LOC_Os10g28640 |
| Inversion | Chr8 | 15462744 | 15596597 | 2 |
| Inversion | Chr8 | 15462759 | 15596597 | 2 |
| Inversion | Chr1 | 18299015 | 18299201 | LOC_Os01g33230 |
| Inversion | Chr10 | 20792513 | 20792750 | LOC_Os10g39010 |
| Inversion | Chr1 | 21352667 | 21352796 | |
| Inversion | Chr12 | 22174310 | 22174487 | LOC_Os12g36180 |
| Inversion | Chr5 | 22236578 | 22236704 | |
| Inversion | Chr4 | 22279901 | 22280086 | |
| Inversion | Chr8 | 22369139 | 22369317 | |
| Inversion | Chr10 | 22435425 | 22435531 | LOC_Os10g41660 |
| Inversion | Chr2 | 22533845 | 22534081 | |
| Inversion | Chr8 | 22816298 | 22816631 | |
| Inversion | Chr6 | 23334923 | 23335081 | LOC_Os06g39330 |
| Inversion | Chr5 | 24155553 | 24155893 | LOC_Os05g41240 |
| Inversion | Chr1 | 25405860 | 25406018 | |
| Inversion | Chr8 | 25614818 | 25615124 | LOC_Os08g40490 |
| Inversion | Chr6 | 26499155 | 26566948 | |
| Inversion | Chr6 | 26499165 | 26566952 | |
| Inversion | Chr3 | 28791267 | 28791681 | |
| Inversion | Chr2 | 29204935 | 29205217 | |
| Inversion | Chr2 | 29611853 | 29612199 | LOC_Os02g48370 |
| Inversion | Chr1 | 34941276 | 35366085 | 2 |
| Inversion | Chr1 | 35366097 | 35721717 | 2 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2785663 | Chr8 | 4650014 | 2 |
| Translocation | Chr8 | 4650007 | Chr2 | 21197285 | LOC_Os08g08170 |
| Translocation | Chr11 | 8192984 | Chr4 | 13912171 | |
| Translocation | Chr10 | 9076083 | Chr4 | 2768659 | |
| Translocation | Chr8 | 12652116 | Chr4 | 4434857 | |
| Translocation | Chr8 | 12662666 | Chr4 | 4434843 | LOC_Os08g21210 |
| Translocation | Chr10 | 18851560 | Chr4 | 34078966 | |
| Translocation | Chr10 | 18851568 | Chr4 | 34071875 | |
| Translocation | Chr2 | 19183034 | Chr1 | 37115681 | |
| Translocation | Chr8 | 19686890 | Chr6 | 13719896 | 2 |
| Translocation | Chr6 | 23462865 | Chr3 | 32007061 | LOC_Os06g39515 |
