Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2196-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2196-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 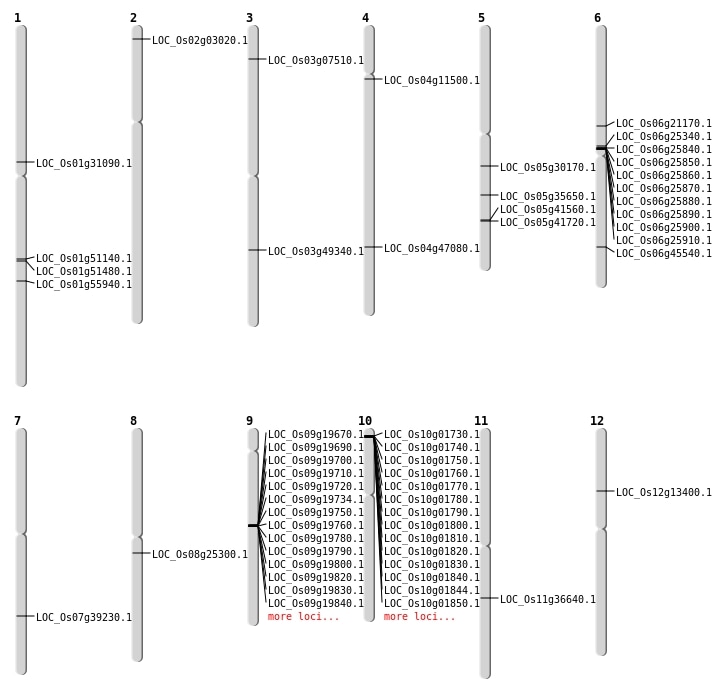
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25171583 | G-A | HET | ||
| SBS | Chr1 | 29587425 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g51480 |
| SBS | Chr1 | 32221477 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g55940 |
| SBS | Chr1 | 32221478 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g55940 |
| SBS | Chr1 | 6364507 | T-A | Homo | ||
| SBS | Chr10 | 10391657 | T-A | HET | ||
| SBS | Chr10 | 11497556 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22220 |
| SBS | Chr10 | 16926941 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g32240 |
| SBS | Chr10 | 4079121 | T-A | Homo | ||
| SBS | Chr10 | 4079122 | G-A | Homo | ||
| SBS | Chr10 | 5978569 | T-A | HET | ||
| SBS | Chr10 | 6486970 | C-A | HET | ||
| SBS | Chr10 | 9238880 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g18190 |
| SBS | Chr11 | 17748869 | C-T | HET | ||
| SBS | Chr11 | 17790171 | T-C | HET | ||
| SBS | Chr11 | 21620269 | C-A | HET | ||
| SBS | Chr11 | 21620270 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36640 |
| SBS | Chr2 | 1187817 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g03020 |
| SBS | Chr2 | 15208476 | G-A | HET | ||
| SBS | Chr2 | 35816681 | T-A | HET | ||
| SBS | Chr2 | 5062641 | A-T | HET | ||
| SBS | Chr2 | 8793772 | C-T | HET | ||
| SBS | Chr3 | 17303087 | A-T | HET | ||
| SBS | Chr3 | 21929921 | A-C | HET | ||
| SBS | Chr3 | 28096542 | A-T | HET | ||
| SBS | Chr3 | 30994779 | C-T | HET | ||
| SBS | Chr3 | 4068329 | G-A | HET | ||
| SBS | Chr4 | 18124292 | G-C | HET | ||
| SBS | Chr4 | 26669462 | C-T | Homo | ||
| SBS | Chr4 | 27954050 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g47080 |
| SBS | Chr4 | 27954051 | C-T | Homo | ||
| SBS | Chr4 | 30452082 | T-C | HET | ||
| SBS | Chr4 | 34189673 | C-A | Homo | ||
| SBS | Chr4 | 3856703 | C-T | Homo | ||
| SBS | Chr4 | 6286717 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g11500 |
| SBS | Chr4 | 7184908 | C-T | HET | ||
| SBS | Chr5 | 17487775 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g30170 |
| SBS | Chr5 | 20601081 | A-T | HET | ||
| SBS | Chr5 | 29302060 | A-G | Homo | ||
| SBS | Chr6 | 27560137 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g45540 |
| SBS | Chr6 | 8423069 | T-A | Homo | ||
| SBS | Chr6 | 8593654 | G-A | Homo | ||
| SBS | Chr7 | 18411595 | A-G | Homo | ||
| SBS | Chr7 | 23487335 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g39230 |
| SBS | Chr7 | 24956842 | C-T | HET | ||
| SBS | Chr7 | 9766931 | C-T | Homo | ||
| SBS | Chr8 | 8402517 | G-A | HET | ||
| SBS | Chr9 | 19088260 | T-A | HET |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 475001 | 677000 | 201999 | 33 |
| Deletion | Chr3 | 741831 | 741838 | 7 | |
| Deletion | Chr5 | 6670092 | 6670096 | 4 | |
| Deletion | Chr5 | 6988359 | 6988360 | 1 | |
| Deletion | Chr5 | 8601605 | 8601606 | 1 | |
| Deletion | Chr6 | 11649304 | 11649310 | 6 | |
| Deletion | Chr9 | 11757001 | 11857000 | 99999 | 11 |
| Deletion | Chr9 | 11862001 | 11946000 | 83999 | 14 |
| Deletion | Chr9 | 11951001 | 11967000 | 15999 | 5 |
| Deletion | Chr9 | 11974001 | 12183000 | 208999 | 31 |
| Deletion | Chr6 | 12235889 | 12235890 | 1 | LOC_Os06g21170 |
| Deletion | Chr9 | 12548925 | 12548929 | 4 | |
| Deletion | Chr6 | 13264184 | 13264188 | 4 | |
| Deletion | Chr3 | 14107086 | 14107088 | 2 | |
| Deletion | Chr3 | 14968565 | 14968579 | 14 | |
| Deletion | Chr6 | 15092001 | 15138000 | 45999 | 5 |
| Deletion | Chr6 | 15142001 | 15166000 | 23999 | 3 |
| Deletion | Chr1 | 16999239 | 16999243 | 4 | LOC_Os01g31090 |
| Deletion | Chr7 | 19358211 | 19358213 | 2 | |
| Deletion | Chr3 | 22596283 | 22596284 | 1 | |
| Deletion | Chr10 | 23040190 | 23040191 | 1 | |
| Deletion | Chr4 | 25597439 | 25597447 | 8 | |
| Deletion | Chr7 | 28978785 | 28978789 | 4 | |
| Deletion | Chr1 | 29384079 | 29384085 | 6 | LOC_Os01g51140 |
| Deletion | Chr2 | 30027405 | 30027417 | 12 | |
| Deletion | Chr1 | 38613786 | 38613788 | 2 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 29926067 | 29926073 | 7 | |
| Insertion | Chr5 | 23858409 | 23858412 | 4 | |
| Insertion | Chr6 | 25426205 | 25426206 | 2 | |
| Insertion | Chr7 | 25931643 | 25931644 | 2 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3821817 | 3822077 | LOC_Os03g07510 |
| Inversion | Chr6 | 14841610 | 15092414 | |
| Inversion | Chr6 | 14842896 | 15169399 | LOC_Os06g25340 |
| Inversion | Chr5 | 21185529 | 21229376 | LOC_Os05g35650 |
| Inversion | Chr5 | 21185537 | 21229376 | LOC_Os05g35650 |
| Inversion | Chr5 | 24339404 | 24411688 | 2 |
| Inversion | Chr5 | 24339453 | 24411693 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 676044 | Chr3 | 28445994 | |
| Translocation | Chr10 | 676729 | Chr9 | 12133652 | 2 |
| Translocation | Chr12 | 7498166 | Chr7 | 13571127 | LOC_Os12g13400 |
| Translocation | Chr9 | 12132382 | Chr4 | 33608212 | |
| Translocation | Chr9 | 12133648 | Chr4 | 33607599 | LOC_Os09g20250 |
| Translocation | Chr9 | 12134682 | Chr3 | 28086646 | 2 |
| Translocation | Chr8 | 15383527 | Chr4 | 7447945 | LOC_Os08g25300 |
