Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2218-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2218-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 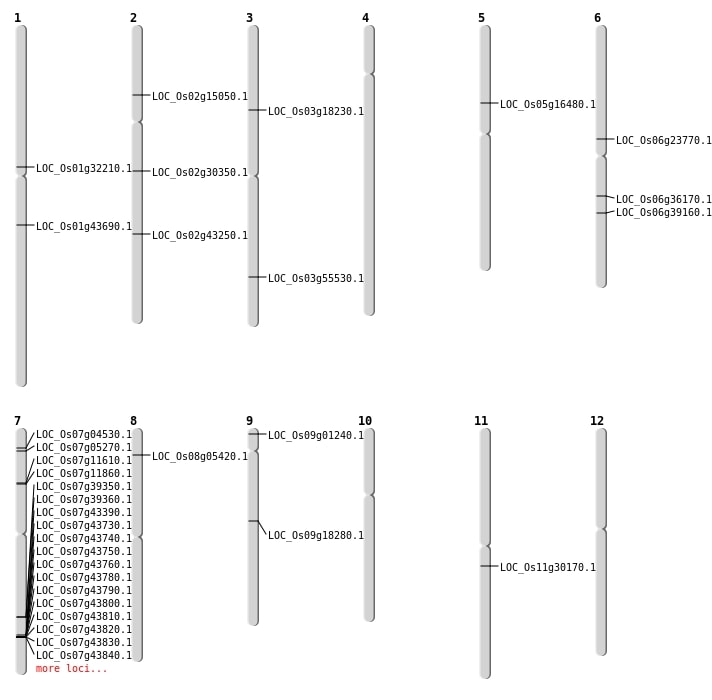
|
| Variant Information |
|---|
Single base substitutions: 80
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15313985 | C-A | HET | ||
| SBS | Chr1 | 19804448 | G-A | HET | ||
| SBS | Chr1 | 21032026 | T-C | HET | ||
| SBS | Chr1 | 28587182 | T-C | HET | ||
| SBS | Chr1 | 4705422 | T-C | HET | ||
| SBS | Chr10 | 10420633 | G-A | HET | ||
| SBS | Chr10 | 8736002 | G-C | HET | ||
| SBS | Chr11 | 16615087 | A-G | Homo | ||
| SBS | Chr11 | 17542950 | C-A | Homo | STOP_GAINED | LOC_Os11g30170 |
| SBS | Chr11 | 8606648 | G-A | HET | ||
| SBS | Chr12 | 15102254 | C-T | HET | ||
| SBS | Chr12 | 2085048 | G-T | HET | ||
| SBS | Chr2 | 10295661 | C-A | HET | ||
| SBS | Chr2 | 10757896 | C-T | HET | ||
| SBS | Chr2 | 17498659 | T-A | HET | ||
| SBS | Chr2 | 20024278 | G-A | HET | ||
| SBS | Chr2 | 21888499 | G-A | HET | ||
| SBS | Chr2 | 26053156 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g43250 |
| SBS | Chr2 | 28603349 | C-T | HET | ||
| SBS | Chr3 | 15543432 | C-A | HET | ||
| SBS | Chr3 | 2139620 | A-G | Homo | ||
| SBS | Chr3 | 22093628 | G-C | Homo | ||
| SBS | Chr3 | 22093632 | A-T | Homo | ||
| SBS | Chr3 | 31597914 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g55530 |
| SBS | Chr3 | 35777781 | A-T | HET | ||
| SBS | Chr3 | 412942 | A-T | Homo | ||
| SBS | Chr3 | 5355301 | G-A | HET | ||
| SBS | Chr4 | 16909327 | G-T | HET | ||
| SBS | Chr4 | 16914119 | G-T | HET | ||
| SBS | Chr4 | 18764248 | C-T | HET | ||
| SBS | Chr4 | 19848142 | C-G | HET | ||
| SBS | Chr4 | 20830707 | T-C | HET | ||
| SBS | Chr4 | 25441191 | C-T | HET | ||
| SBS | Chr4 | 27356244 | T-A | HET | ||
| SBS | Chr4 | 28445423 | T-C | HET | ||
| SBS | Chr4 | 29763956 | C-A | HET | ||
| SBS | Chr5 | 14050171 | A-G | Homo | ||
| SBS | Chr5 | 1761646 | C-T | HET | ||
| SBS | Chr5 | 4286656 | G-T | HET | ||
| SBS | Chr5 | 9354726 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16480 |
| SBS | Chr6 | 1177122 | G-A | HET | ||
| SBS | Chr6 | 1177129 | G-A | HET | ||
| SBS | Chr6 | 13886017 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23770 |
| SBS | Chr6 | 15912545 | C-T | Homo | ||
| SBS | Chr6 | 21171252 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g36170 |
| SBS | Chr6 | 23307621 | G-T | HET | ||
| SBS | Chr6 | 23834322 | C-A | HET | ||
| SBS | Chr6 | 25220246 | T-C | HET | ||
| SBS | Chr6 | 29683841 | G-T | HET | ||
| SBS | Chr6 | 702389 | G-T | HET | ||
| SBS | Chr7 | 14573845 | A-G | HET | ||
| SBS | Chr7 | 14615977 | A-C | HET | ||
| SBS | Chr7 | 20453759 | A-T | Homo | ||
| SBS | Chr7 | 2377677 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g05270 |
| SBS | Chr7 | 25844572 | A-T | HET | ||
| SBS | Chr7 | 26197324 | C-T | Homo | ||
| SBS | Chr7 | 26790483 | C-T | HET | ||
| SBS | Chr7 | 27576931 | A-T | HET | ||
| SBS | Chr7 | 28697034 | A-G | HET | ||
| SBS | Chr7 | 28805464 | G-A | HET | ||
| SBS | Chr7 | 28956813 | C-T | HET | ||
| SBS | Chr7 | 29495158 | C-T | HET | ||
| SBS | Chr7 | 6407089 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11610 |
| SBS | Chr7 | 6564155 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11860 |
| SBS | Chr7 | 7075868 | T-A | HET | ||
| SBS | Chr7 | 7075881 | T-A | HET | ||
| SBS | Chr7 | 8653587 | C-G | HET | ||
| SBS | Chr7 | 9481352 | G-T | HET | ||
| SBS | Chr7 | 9944147 | G-A | HET | ||
| SBS | Chr7 | 9944834 | G-T | HET | ||
| SBS | Chr8 | 10084903 | C-A | Homo | ||
| SBS | Chr8 | 16143536 | T-A | Homo | ||
| SBS | Chr8 | 19239322 | C-A | HET | ||
| SBS | Chr8 | 19624639 | C-T | Homo | ||
| SBS | Chr8 | 3763188 | C-A | HET | ||
| SBS | Chr8 | 7182772 | C-A | HET | ||
| SBS | Chr8 | 8144557 | G-A | HET | ||
| SBS | Chr9 | 11219819 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18280 |
| SBS | Chr9 | 4770574 | A-G | Homo | ||
| SBS | Chr9 | 5670778 | G-A | Homo |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2002556 | 2002577 | 21 | LOC_Os07g04530 |
| Deletion | Chr11 | 2378625 | 2378626 | 1 | |
| Deletion | Chr10 | 4005287 | 4005289 | 2 | |
| Deletion | Chr11 | 5658136 | 5658141 | 5 | |
| Deletion | Chr2 | 8395115 | 8395121 | 6 | LOC_Os02g15050 |
| Deletion | Chr3 | 11249344 | 11249355 | 11 | |
| Deletion | Chr5 | 12102798 | 12102799 | 1 | |
| Deletion | Chr5 | 14971763 | 14971766 | 3 | |
| Deletion | Chr10 | 15235739 | 15235743 | 4 | |
| Deletion | Chr12 | 16489282 | 16489283 | 1 | |
| Deletion | Chr3 | 16566912 | 16566913 | 1 | |
| Deletion | Chr1 | 16637544 | 16637545 | 1 | |
| Deletion | Chr4 | 21532558 | 21532560 | 2 | |
| Deletion | Chr5 | 24342370 | 24342372 | 2 | |
| Deletion | Chr7 | 26171001 | 26521000 | 349999 | 63 |
| Deletion | Chr2 | 30414705 | 30414706 | 1 | |
| Deletion | Chr4 | 31962421 | 31962422 | 1 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 24275651 | 24275652 | 2 | |
| Insertion | Chr3 | 10224923 | 10224924 | 2 | LOC_Os03g18230 |
| Insertion | Chr4 | 4268572 | 4268572 | 1 | |
| Insertion | Chr8 | 22554866 | 22554867 | 2 | |
| Insertion | Chr9 | 3843609 | 3843611 | 3 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 25740548 | 25845783 | |
| Inversion | Chr7 | 25753275 | 25983225 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 202840 | Chr6 | 14437353 | LOC_Os09g01240 |
| Translocation | Chr9 | 202853 | Chr6 | 14437164 | LOC_Os09g01240 |
| Translocation | Chr8 | 2861073 | Chr1 | 17634409 | 2 |
| Translocation | Chr6 | 7292234 | Chr2 | 18076972 | 2 |
| Translocation | Chr7 | 22897469 | Chr3 | 11855839 | |
| Translocation | Chr6 | 23251200 | Chr3 | 13645054 | LOC_Os06g39160 |
| Translocation | Chr7 | 23583901 | Chr1 | 25038571 | 2 |
| Translocation | Chr7 | 23585458 | Chr1 | 25038602 | 2 |
