Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2274-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2274-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 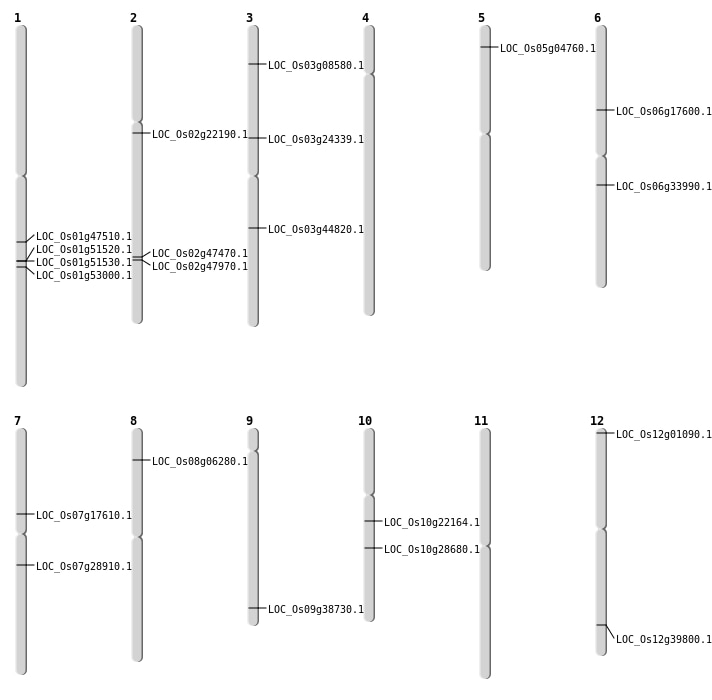
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14308741 | T-C | HET | ||
| SBS | Chr1 | 27157074 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g47510 |
| SBS | Chr1 | 34072195 | G-A | HET | ||
| SBS | Chr1 | 35076680 | G-A | HET | ||
| SBS | Chr1 | 40685653 | T-A | Homo | ||
| SBS | Chr1 | 41568831 | G-T | HET | ||
| SBS | Chr1 | 41728049 | G-A | HET | ||
| SBS | Chr1 | 9716014 | C-G | HET | ||
| SBS | Chr10 | 11458867 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g22164 |
| SBS | Chr10 | 17421836 | T-C | HET | ||
| SBS | Chr10 | 17769049 | A-G | HET | ||
| SBS | Chr10 | 17769196 | T-G | HET | ||
| SBS | Chr10 | 5486005 | T-A | Homo | ||
| SBS | Chr10 | 5627772 | G-A | HET | ||
| SBS | Chr11 | 20370010 | T-A | Homo | ||
| SBS | Chr12 | 14668462 | C-T | HET | ||
| SBS | Chr12 | 2270893 | G-A | HET | ||
| SBS | Chr12 | 47560 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g01090 |
| SBS | Chr12 | 506079 | C-T | HET | ||
| SBS | Chr12 | 7789171 | C-T | HET | ||
| SBS | Chr2 | 11736824 | A-G | HET | ||
| SBS | Chr2 | 17198258 | A-T | HET | ||
| SBS | Chr2 | 23147050 | C-T | HET | ||
| SBS | Chr2 | 27832725 | G-A | HET | ||
| SBS | Chr2 | 34569832 | G-A | Homo | ||
| SBS | Chr2 | 5825965 | A-G | HET | ||
| SBS | Chr3 | 25277321 | C-A | HET | ||
| SBS | Chr3 | 25277322 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g44820 |
| SBS | Chr3 | 4417197 | T-C | Homo | SPLICE_SITE_DONOR | LOC_Os03g08580 |
| SBS | Chr3 | 8439732 | T-C | HET | ||
| SBS | Chr3 | 9357165 | G-A | Homo | ||
| SBS | Chr4 | 15346902 | G-C | HET | ||
| SBS | Chr4 | 1538725 | C-T | Homo | ||
| SBS | Chr4 | 15548564 | T-A | HET | ||
| SBS | Chr4 | 7676544 | C-T | HET | ||
| SBS | Chr5 | 21064383 | A-G | HET | ||
| SBS | Chr5 | 28255715 | G-C | HET | ||
| SBS | Chr5 | 5598503 | C-T | HET | ||
| SBS | Chr5 | 7862297 | A-T | Homo | ||
| SBS | Chr6 | 10213883 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g17600 |
| SBS | Chr6 | 10213884 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g17600 |
| SBS | Chr6 | 11367176 | G-C | Homo | ||
| SBS | Chr6 | 23825898 | G-A | HET | ||
| SBS | Chr7 | 10402257 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17610 |
| SBS | Chr7 | 10413177 | G-T | HET | ||
| SBS | Chr7 | 17571023 | A-T | HET | ||
| SBS | Chr8 | 13109746 | T-C | HET | ||
| SBS | Chr8 | 14911077 | G-A | HET | ||
| SBS | Chr8 | 20901584 | G-A | HET | ||
| SBS | Chr8 | 3701036 | A-T | Homo | ||
| SBS | Chr9 | 15433984 | A-G | HET |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1487284 | 1487285 | 1 | |
| Deletion | Chr10 | 2161262 | 2161265 | 3 | |
| Deletion | Chr5 | 2253871 | 2253881 | 10 | LOC_Os05g04760 |
| Deletion | Chr8 | 2818922 | 2818924 | 2 | |
| Deletion | Chr9 | 3409287 | 3409289 | 2 | |
| Deletion | Chr8 | 4016292 | 4016302 | 10 | |
| Deletion | Chr1 | 4448250 | 4448254 | 4 | |
| Deletion | Chr8 | 4480183 | 4480187 | 4 | |
| Deletion | Chr2 | 6824165 | 6824167 | 2 | |
| Deletion | Chr10 | 9625833 | 9625835 | 2 | |
| Deletion | Chr3 | 11041306 | 11041310 | 4 | |
| Deletion | Chr3 | 12272208 | 12272209 | 1 | |
| Deletion | Chr3 | 13841597 | 13841606 | 9 | LOC_Os03g24339 |
| Deletion | Chr10 | 14962123 | 14962125 | 2 | LOC_Os10g28680 |
| Deletion | Chr4 | 15866142 | 15866143 | 1 | |
| Deletion | Chr10 | 16709703 | 16709747 | 44 | |
| Deletion | Chr3 | 20543498 | 20543508 | 10 | |
| Deletion | Chr6 | 21695000 | 21695001 | 1 | |
| Deletion | Chr11 | 24168436 | 24168441 | 5 | |
| Deletion | Chr6 | 25465865 | 25465887 | 22 | |
| Deletion | Chr2 | 26774530 | 26774535 | 5 | |
| Deletion | Chr1 | 29596001 | 29605000 | 8999 | 2 |
| Deletion | Chr6 | 30141157 | 30141166 | 9 | |
| Deletion | Chr1 | 42137413 | 42137418 | 5 | |
| Deletion | Chr1 | 42166637 | 42166639 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr8 | 17546054 | 17546055 | 2 | |
| Insertion | Chr8 | 5130577 | 5130578 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 21930722 | 22261288 | 2 |
| Inversion | Chr2 | 28701309 | 28995921 | 2 |
| Inversion | Chr2 | 28995922 | 29348217 | 2 |
| Inversion | Chr1 | 29595676 | 30461571 | 2 |
| Inversion | Chr1 | 29604731 | 30461578 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 728626 | Chr2 | 13225538 | 2 |
| Translocation | Chr8 | 3463065 | Chr7 | 16953256 | 2 |
| Translocation | Chr12 | 24587182 | Chr6 | 19795359 | 2 |
