Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2275-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2275-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 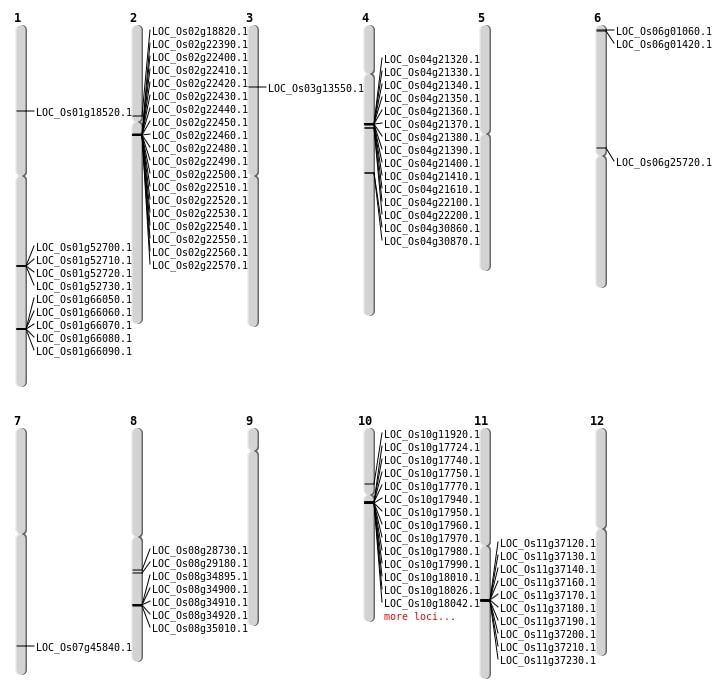
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2557267 | T-C | HET | ||
| SBS | Chr10 | 16849900 | C-T | HET | ||
| SBS | Chr10 | 19807880 | G-T | HET | ||
| SBS | Chr10 | 6082314 | A-T | HET | ||
| SBS | Chr10 | 6176012 | G-T | HET | ||
| SBS | Chr10 | 8391705 | A-T | HET | ||
| SBS | Chr10 | 9539422 | G-A | HET | ||
| SBS | Chr10 | 9679536 | T-A | HET | ||
| SBS | Chr11 | 11644498 | T-A | HET | ||
| SBS | Chr11 | 14261627 | G-A | HET | ||
| SBS | Chr11 | 3937732 | G-A | HET | ||
| SBS | Chr11 | 5821433 | G-A | Homo | ||
| SBS | Chr12 | 10800352 | T-C | HET | ||
| SBS | Chr12 | 16771263 | G-C | Homo | ||
| SBS | Chr2 | 10980386 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18820 |
| SBS | Chr2 | 15008774 | T-A | HET | ||
| SBS | Chr2 | 15009193 | C-A | HET | ||
| SBS | Chr2 | 15009194 | G-A | HET | ||
| SBS | Chr2 | 15009195 | C-T | HET | ||
| SBS | Chr2 | 28145108 | T-A | Homo | ||
| SBS | Chr2 | 34686306 | G-A | HET | ||
| SBS | Chr3 | 11982782 | A-G | HET | ||
| SBS | Chr3 | 1508600 | G-A | Homo | ||
| SBS | Chr3 | 19696004 | C-T | Homo | ||
| SBS | Chr3 | 7326501 | T-A | Homo | STOP_GAINED | LOC_Os03g13550 |
| SBS | Chr4 | 10313619 | G-A | HET | ||
| SBS | Chr4 | 15414853 | T-C | HET | ||
| SBS | Chr4 | 17413893 | C-T | HET | ||
| SBS | Chr4 | 17456279 | T-A | HET | ||
| SBS | Chr4 | 29336760 | T-G | HET | ||
| SBS | Chr5 | 5769934 | C-T | Homo | ||
| SBS | Chr6 | 15020809 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g25720 |
| SBS | Chr6 | 2281446 | T-G | HET | ||
| SBS | Chr7 | 27358506 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g45840 |
| SBS | Chr8 | 11002447 | A-T | HET | ||
| SBS | Chr8 | 11131788 | G-A | HET | ||
| SBS | Chr8 | 4923838 | C-T | HET | ||
| SBS | Chr9 | 5627103 | T-A | HET |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 859946 | 859947 | 1 | |
| Deletion | Chr2 | 957683 | 957686 | 3 | |
| Deletion | Chr2 | 2059555 | 2059557 | 2 | |
| Deletion | Chr10 | 5832710 | 5832711 | 1 | |
| Deletion | Chr10 | 6615820 | 6615823 | 3 | LOC_Os10g11920 |
| Deletion | Chr6 | 7810371 | 7810379 | 8 | |
| Deletion | Chr12 | 8022335 | 8022350 | 15 | |
| Deletion | Chr12 | 8699756 | 8699761 | 5 | |
| Deletion | Chr10 | 8962001 | 8995000 | 32999 | 4 |
| Deletion | Chr10 | 9071001 | 9172000 | 100999 | 14 |
| Deletion | Chr4 | 12023001 | 12104000 | 80999 | 10 |
| Deletion | Chr8 | 12655551 | 12655554 | 3 | |
| Deletion | Chr9 | 13068656 | 13068665 | 9 | |
| Deletion | Chr2 | 13374001 | 13398000 | 23999 | 5 |
| Deletion | Chr2 | 13404001 | 13461000 | 56999 | 13 |
| Deletion | Chr7 | 14568224 | 14568229 | 5 | |
| Deletion | Chr11 | 15784490 | 15784491 | 1 | |
| Deletion | Chr10 | 17747083 | 17747089 | 6 | |
| Deletion | Chr4 | 17819989 | 17819990 | 1 | |
| Deletion | Chr4 | 18442001 | 18450000 | 7999 | 2 |
| Deletion | Chr9 | 18903572 | 18903576 | 4 | |
| Deletion | Chr11 | 21915001 | 21993000 | 77999 | 10 |
| Deletion | Chr8 | 21973001 | 21991000 | 17999 | 4 |
| Deletion | Chr8 | 22054558 | 22054563 | 5 | LOC_Os08g35010 |
| Deletion | Chr8 | 23802263 | 23802264 | 1 | |
| Deletion | Chr11 | 24292964 | 24292966 | 2 | |
| Deletion | Chr11 | 25183222 | 25183224 | 2 | |
| Deletion | Chr8 | 26226637 | 26226639 | 2 | |
| Deletion | Chr12 | 26812100 | 26812131 | 31 | |
| Deletion | Chr1 | 30298001 | 30330000 | 31999 | 4 |
| Deletion | Chr2 | 32293947 | 32293955 | 8 | |
| Deletion | Chr3 | 33695951 | 33695954 | 3 | |
| Deletion | Chr2 | 33866118 | 33866120 | 2 | |
| Deletion | Chr1 | 38331001 | 38367000 | 35999 | 5 |
| Deletion | Chr1 | 39023200 | 39023206 | 6 | |
| Deletion | Chr1 | 41689716 | 41689719 | 3 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10418465 | 10418465 | 1 | LOC_Os01g18520 |
| Insertion | Chr1 | 10882620 | 10882622 | 3 | |
| Insertion | Chr10 | 20202591 | 20202592 | 2 | |
| Insertion | Chr11 | 9368836 | 9368838 | 3 | |
| Insertion | Chr2 | 20960937 | 20960938 | 2 | |
| Insertion | Chr2 | 33270300 | 33270302 | 3 | |
| Insertion | Chr7 | 6252890 | 6252891 | 2 | |
| Insertion | Chr8 | 18175699 | 18175702 | 4 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 46338 | 255774 | 2 |
| Inversion | Chr10 | 8992716 | 9169360 | LOC_Os10g17770 |
| Inversion | Chr4 | 12520367 | 12576935 | 2 |
| Inversion | Chr6 | 17296350 | 17296648 | |
| Inversion | Chr8 | 17574107 | 17883151 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 14462355 | Chr4 | 12223961 | 2 |
| Translocation | Chr12 | 14462356 | Chr4 | 12222064 | 2 |
