Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2279-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2279-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 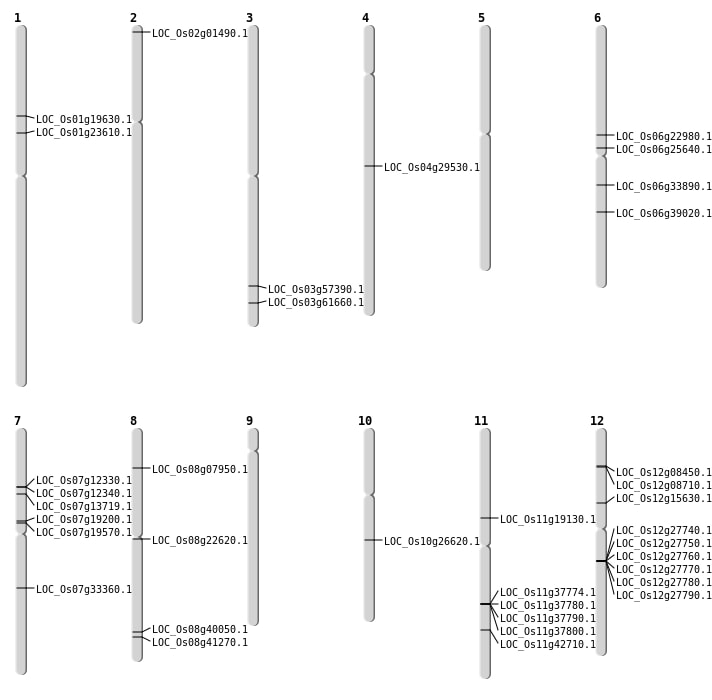
|
| Variant Information |
|---|
Single base substitutions: 62
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11129884 | C-T | HET | STOP_GAINED | LOC_Os01g19630 |
| SBS | Chr1 | 20256603 | T-C | HET | ||
| SBS | Chr1 | 22051332 | C-T | Homo | ||
| SBS | Chr1 | 23097603 | A-C | Homo | ||
| SBS | Chr1 | 23097608 | C-T | Homo | ||
| SBS | Chr1 | 26749343 | C-T | HET | ||
| SBS | Chr1 | 34586345 | G-A | HET | ||
| SBS | Chr10 | 13860456 | A-T | Homo | ||
| SBS | Chr10 | 1395228 | A-C | Homo | ||
| SBS | Chr10 | 16833793 | A-C | HET | ||
| SBS | Chr10 | 20329618 | T-C | HET | ||
| SBS | Chr10 | 5175766 | C-G | HET | ||
| SBS | Chr11 | 17876829 | C-T | HET | ||
| SBS | Chr11 | 18408479 | G-A | HET | ||
| SBS | Chr11 | 18456697 | T-G | HET | ||
| SBS | Chr11 | 217467 | T-C | HET | ||
| SBS | Chr11 | 6303871 | C-T | HET | ||
| SBS | Chr11 | 6303872 | C-T | HET | ||
| SBS | Chr11 | 778173 | C-T | Homo | ||
| SBS | Chr12 | 10976062 | T-A | HET | ||
| SBS | Chr12 | 10986986 | C-T | HET | ||
| SBS | Chr12 | 147021 | C-T | Homo | ||
| SBS | Chr12 | 8928036 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15630 |
| SBS | Chr2 | 2585902 | C-T | Homo | ||
| SBS | Chr2 | 2585903 | G-T | Homo | ||
| SBS | Chr3 | 16309531 | C-T | HET | ||
| SBS | Chr3 | 24425352 | A-T | HET | ||
| SBS | Chr3 | 30215260 | T-C | HET | ||
| SBS | Chr4 | 18422998 | G-A | HET | ||
| SBS | Chr4 | 19938193 | G-T | HET | ||
| SBS | Chr4 | 24825895 | C-T | Homo | ||
| SBS | Chr4 | 27339915 | A-G | HET | ||
| SBS | Chr5 | 12122345 | C-T | Homo | ||
| SBS | Chr5 | 14041055 | T-A | Homo | ||
| SBS | Chr5 | 20494055 | C-T | HET | ||
| SBS | Chr5 | 21336043 | T-C | Homo | ||
| SBS | Chr5 | 7611148 | G-A | HET | ||
| SBS | Chr6 | 10103741 | T-C | HET | ||
| SBS | Chr6 | 13234457 | G-A | HET | ||
| SBS | Chr6 | 13416046 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g22980 |
| SBS | Chr6 | 14988154 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25640 |
| SBS | Chr6 | 15736816 | C-T | HET | ||
| SBS | Chr6 | 19717550 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g33890 |
| SBS | Chr6 | 23166144 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g39020 |
| SBS | Chr6 | 23166160 | G-A | HET | ||
| SBS | Chr6 | 2719032 | G-T | HET | ||
| SBS | Chr6 | 27465124 | C-T | HET | ||
| SBS | Chr6 | 30822879 | G-A | HET | ||
| SBS | Chr6 | 6959951 | C-T | Homo | ||
| SBS | Chr7 | 11598160 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 19941630 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g33360 |
| SBS | Chr7 | 24259100 | T-C | Homo | ||
| SBS | Chr8 | 13609442 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g22620 |
| SBS | Chr8 | 18808807 | C-T | Homo | ||
| SBS | Chr8 | 19172575 | C-T | HET | ||
| SBS | Chr8 | 20660875 | G-T | HET | ||
| SBS | Chr8 | 26066163 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g41270 |
| SBS | Chr8 | 3454627 | A-T | Homo | ||
| SBS | Chr8 | 9191549 | C-T | Homo | ||
| SBS | Chr9 | 17469763 | G-A | Homo | ||
| SBS | Chr9 | 17469764 | G-C | Homo | ||
| SBS | Chr9 | 21019259 | C-T | Homo |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 844605 | 844613 | 8 | |
| Deletion | Chr8 | 1727982 | 1727987 | 5 | |
| Deletion | Chr2 | 2999868 | 2999881 | 13 | |
| Deletion | Chr6 | 4429682 | 4429683 | 1 | |
| Deletion | Chr1 | 6381555 | 6381566 | 11 | |
| Deletion | Chr7 | 6959001 | 6988000 | 28999 | 2 |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr6 | 10419568 | 10419571 | 3 | |
| Deletion | Chr4 | 11579539 | 11579543 | 4 | |
| Deletion | Chr5 | 11725086 | 11725095 | 9 | |
| Deletion | Chr11 | 13231450 | 13231453 | 3 | |
| Deletion | Chr1 | 13277001 | 13283000 | 5999 | LOC_Os01g23610 |
| Deletion | Chr12 | 13628389 | 13628395 | 6 | |
| Deletion | Chr10 | 13882001 | 13891000 | 8999 | LOC_Os10g26620 |
| Deletion | Chr10 | 15484871 | 15484873 | 2 | |
| Deletion | Chr12 | 16357001 | 16384000 | 26999 | 6 |
| Deletion | Chr9 | 18290035 | 18290036 | 1 | |
| Deletion | Chr5 | 19279454 | 19279455 | 1 | |
| Deletion | Chr9 | 20204810 | 20204814 | 4 | |
| Deletion | Chr2 | 20394886 | 20394892 | 6 | |
| Deletion | Chr8 | 20998792 | 20998809 | 17 | |
| Deletion | Chr11 | 22342001 | 22393000 | 50999 | 4 |
| Deletion | Chr6 | 23150806 | 23150807 | 1 | |
| Deletion | Chr11 | 25727919 | 25727922 | 3 | LOC_Os11g42710 |
| Deletion | Chr1 | 31854812 | 31854814 | 2 | |
| Deletion | Chr3 | 32731431 | 32731432 | 1 | LOC_Os03g57390 |
| Deletion | Chr3 | 34948540 | 34948544 | 4 | LOC_Os03g61660 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 24126767 | 24126768 | 2 | |
| Insertion | Chr1 | 37140758 | 37140758 | 1 | |
| Insertion | Chr3 | 14316094 | 14316095 | 2 | |
| Insertion | Chr8 | 25361005 | 25361005 | 1 | LOC_Os08g40050 |
| Insertion | Chr8 | 2871634 | 2871635 | 2 | |
| Insertion | Chr8 | 3347906 | 3347907 | 2 |
Inversions: 7
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4295588 | Chr7 | 7885934 | LOC_Os12g08450 |
| Translocation | Chr12 | 4435112 | Chr7 | 7875864 | 2 |
| Translocation | Chr6 | 9794993 | Chr4 | 17545681 | 2 |
| Translocation | Chr11 | 10912014 | Chr8 | 4491757 | 2 |
| Translocation | Chr11 | 10912037 | Chr8 | 4465398 | LOC_Os11g19130 |
