Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2485 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2485 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 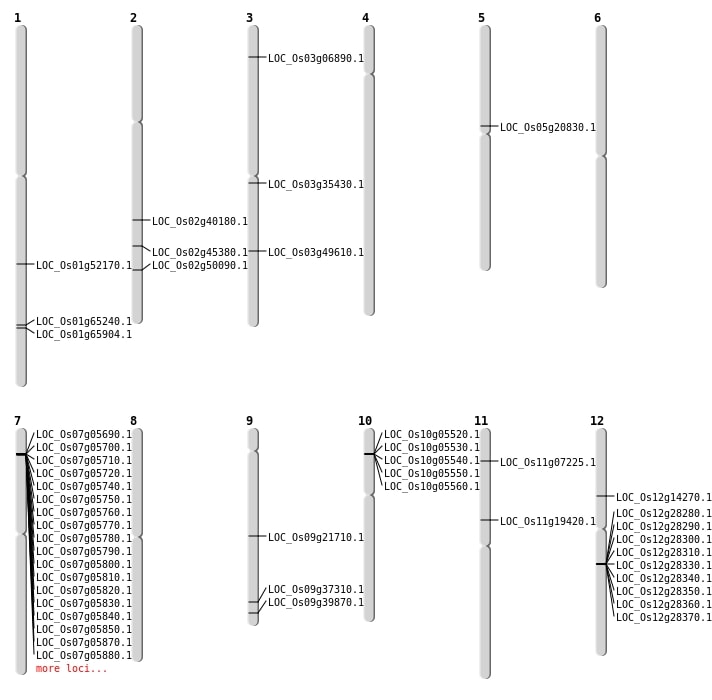
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21088552 | C-A | HET | ||
| SBS | Chr1 | 27704866 | G-C | HET | ||
| SBS | Chr1 | 30010551 | G-A | HET | ||
| SBS | Chr1 | 30010552 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g52170 |
| SBS | Chr1 | 36211166 | A-G | HET | ||
| SBS | Chr1 | 3719129 | A-G | HET | ||
| SBS | Chr1 | 38274510 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g65904 |
| SBS | Chr10 | 3383180 | T-A | Homo | ||
| SBS | Chr10 | 5324440 | G-A | Homo | ||
| SBS | Chr11 | 16749525 | G-A | Homo | ||
| SBS | Chr11 | 3819558 | T-A | Homo | ||
| SBS | Chr12 | 17405505 | T-C | Homo | ||
| SBS | Chr12 | 8122896 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14270 |
| SBS | Chr2 | 11557591 | C-A | HET | ||
| SBS | Chr2 | 27205565 | T-A | Homo | ||
| SBS | Chr2 | 28727578 | G-T | Homo | ||
| SBS | Chr2 | 30588927 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g50090 |
| SBS | Chr2 | 5617126 | G-A | Homo | ||
| SBS | Chr3 | 11988939 | G-A | Homo | ||
| SBS | Chr3 | 19644427 | T-A | Homo | STOP_GAINED | LOC_Os03g35430 |
| SBS | Chr3 | 30371667 | C-G | HET | ||
| SBS | Chr3 | 3485630 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g06890 |
| SBS | Chr3 | 3485632 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g06890 |
| SBS | Chr4 | 13059975 | T-G | HET | ||
| SBS | Chr4 | 24179144 | A-T | HET | ||
| SBS | Chr4 | 2543333 | A-G | HET | ||
| SBS | Chr4 | 26538894 | C-T | HET | ||
| SBS | Chr4 | 31585635 | T-A | HET | ||
| SBS | Chr4 | 6208089 | G-T | HET | ||
| SBS | Chr5 | 10130084 | C-T | Homo | ||
| SBS | Chr5 | 12232953 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20830 |
| SBS | Chr5 | 19793092 | T-C | Homo | ||
| SBS | Chr5 | 21268343 | C-A | HET | ||
| SBS | Chr6 | 12961714 | C-T | HET | ||
| SBS | Chr6 | 17263294 | T-A | HET | ||
| SBS | Chr6 | 30350473 | G-A | HET | ||
| SBS | Chr6 | 4804024 | G-T | HET | ||
| SBS | Chr6 | 9221431 | A-G | HET | ||
| SBS | Chr7 | 12747002 | T-C | HET | ||
| SBS | Chr7 | 13180431 | T-C | HET | ||
| SBS | Chr7 | 23855936 | G-A | Homo | ||
| SBS | Chr7 | 3287861 | G-A | HET | ||
| SBS | Chr8 | 17948674 | A-T | HET | ||
| SBS | Chr8 | 18565123 | C-A | Homo | ||
| SBS | Chr8 | 27341583 | A-T | HET | ||
| SBS | Chr9 | 11247129 | C-T | HET | ||
| SBS | Chr9 | 12264738 | T-C | HET | ||
| SBS | Chr9 | 17664849 | G-A | HET | ||
| SBS | Chr9 | 20671038 | T-A | HET | ||
| SBS | Chr9 | 2286908 | C-T | HET | ||
| SBS | Chr9 | 6312484 | C-T | Homo | ||
| SBS | Chr9 | 9070708 | G-T | HET | ||
| SBS | Chr9 | 9677571 | T-C | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2719001 | 2917000 | 197999 | 31 |
| Deletion | Chr10 | 2732001 | 2755000 | 22999 | 4 |
| Deletion | Chr6 | 4641327 | 4641331 | 4 | |
| Deletion | Chr5 | 8713845 | 8713849 | 4 | |
| Deletion | Chr5 | 10916162 | 10916163 | 1 | |
| Deletion | Chr11 | 11165665 | 11165671 | 6 | LOC_Os11g19420 |
| Deletion | Chr9 | 13155528 | 13155529 | 1 | LOC_Os09g21710 |
| Deletion | Chr7 | 13875001 | 13907000 | 31999 | 5 |
| Deletion | Chr7 | 14767001 | 14777000 | 9999 | 2 |
| Deletion | Chr12 | 16714001 | 16781000 | 66999 | 9 |
| Deletion | Chr8 | 17948281 | 17948282 | 1 | |
| Deletion | Chr5 | 19326027 | 19326028 | 1 | |
| Deletion | Chr5 | 21772839 | 21772840 | 1 | |
| Deletion | Chr9 | 22856146 | 22856164 | 18 | LOC_Os09g39870 |
| Deletion | Chr12 | 23455244 | 23455246 | 2 | |
| Deletion | Chr2 | 27204171 | 27204179 | 8 | |
| Deletion | Chr2 | 27398871 | 27398883 | 12 | |
| Deletion | Chr2 | 27591557 | 27591560 | 3 | LOC_Os02g45380 |
| Deletion | Chr3 | 28252397 | 28252398 | 1 | LOC_Os03g49610 |
| Deletion | Chr3 | 36141704 | 36141706 | 2 | |
| Deletion | Chr1 | 37861161 | 37861162 | 1 | LOC_Os01g65240 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2761381 | 2762165 | LOC_Os10g05560 |
| Inversion | Chr7 | 19100873 | 20069195 | LOC_Os07g32170 |
| Inversion | Chr7 | 19100898 | 20069198 | LOC_Os07g32170 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2239005 | Chr1 | 2819544 | |
| Translocation | Chr11 | 3630102 | Chr2 | 24318498 | 2 |
| Translocation | Chr9 | 8406220 | Chr6 | 11805215 | |
| Translocation | Chr9 | 21552287 | Chr2 | 34253056 | LOC_Os09g37310 |
| Translocation | Chr11 | 22131471 | Chr7 | 11905179 | |
| Translocation | Chr12 | 23684507 | Chr1 | 14833841 |
