Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2531-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2531-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 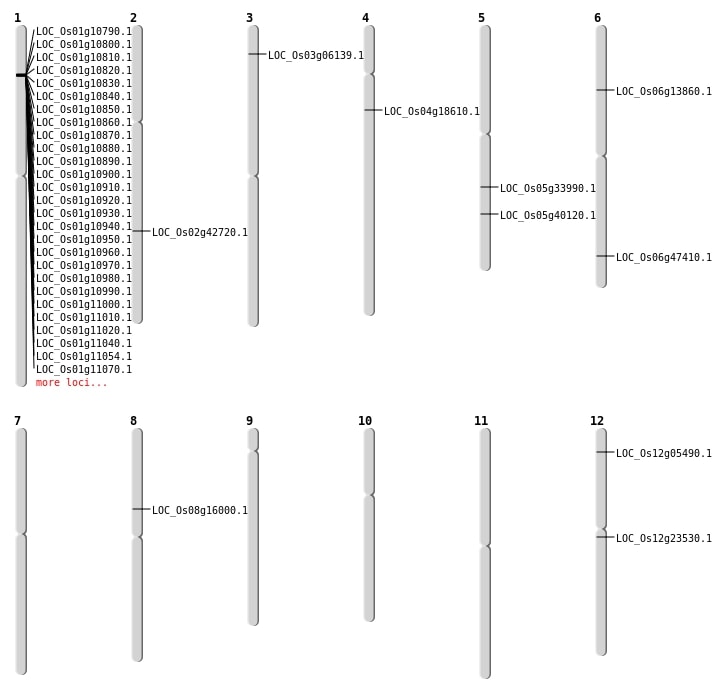
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13987804 | T-A | Homo | ||
| SBS | Chr1 | 17894348 | T-C | HET | ||
| SBS | Chr1 | 19472528 | G-T | HET | ||
| SBS | Chr1 | 21781736 | C-T | HET | ||
| SBS | Chr1 | 42444535 | A-T | HET | ||
| SBS | Chr1 | 9367653 | G-C | HET | ||
| SBS | Chr10 | 5946938 | T-A | HET | ||
| SBS | Chr10 | 8620937 | C-A | HET | ||
| SBS | Chr11 | 17294588 | T-C | HET | ||
| SBS | Chr11 | 18801668 | G-A | HET | ||
| SBS | Chr12 | 10333541 | C-A | HET | ||
| SBS | Chr12 | 13317760 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23530 |
| SBS | Chr12 | 16232356 | C-T | HET | ||
| SBS | Chr12 | 20683832 | T-C | HET | ||
| SBS | Chr12 | 23417069 | C-A | HET | ||
| SBS | Chr12 | 4656568 | T-A | HET | ||
| SBS | Chr12 | 4656569 | T-A | HET | ||
| SBS | Chr12 | 7225046 | C-T | HET | ||
| SBS | Chr2 | 26007086 | C-T | HET | ||
| SBS | Chr2 | 28011188 | G-A | HET | ||
| SBS | Chr2 | 30953152 | T-G | HET | ||
| SBS | Chr2 | 31476248 | A-T | HET | ||
| SBS | Chr2 | 35260751 | T-C | HET | ||
| SBS | Chr3 | 19005267 | C-G | HET | ||
| SBS | Chr3 | 24571688 | C-T | Homo | ||
| SBS | Chr3 | 3076618 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g06139 |
| SBS | Chr3 | 3076619 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g06139 |
| SBS | Chr3 | 4941281 | C-T | HET | ||
| SBS | Chr4 | 10292766 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g18610 |
| SBS | Chr4 | 18295007 | G-A | HET | ||
| SBS | Chr4 | 24972508 | G-T | HET | ||
| SBS | Chr4 | 2769758 | T-C | HET | ||
| SBS | Chr4 | 29327638 | G-A | HET | ||
| SBS | Chr4 | 33361832 | C-A | HET | ||
| SBS | Chr5 | 18054708 | T-A | HET | ||
| SBS | Chr5 | 20069064 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g33990 |
| SBS | Chr5 | 23572057 | A-T | Homo | STOP_GAINED | LOC_Os05g40120 |
| SBS | Chr5 | 6586610 | C-T | HET | ||
| SBS | Chr6 | 11035476 | A-G | HET | ||
| SBS | Chr6 | 20447878 | C-T | HET | ||
| SBS | Chr6 | 23321809 | G-A | HET | ||
| SBS | Chr6 | 28725432 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g47410 |
| SBS | Chr6 | 5562925 | T-C | HET | ||
| SBS | Chr6 | 7822655 | A-G | HET | ||
| SBS | Chr7 | 20966750 | T-A | HET | ||
| SBS | Chr7 | 23263442 | T-C | HET | ||
| SBS | Chr7 | 26804185 | G-T | Homo | ||
| SBS | Chr7 | 4716089 | C-G | Homo | ||
| SBS | Chr8 | 5484824 | T-A | HET | ||
| SBS | Chr8 | 9747302 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g16000 |
| SBS | Chr9 | 1532733 | G-A | HET | ||
| SBS | Chr9 | 6318875 | T-C | HET |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2511522 | 2511529 | 7 | LOC_Os12g05490 |
| Deletion | Chr1 | 5764001 | 6162000 | 397999 | 57 |
| Deletion | Chr7 | 6154911 | 6154933 | 22 | |
| Deletion | Chr1 | 6166001 | 6276000 | 109999 | 14 |
| Deletion | Chr11 | 8582340 | 8582341 | 1 | |
| Deletion | Chr10 | 10388585 | 10388595 | 10 | |
| Deletion | Chr1 | 13435001 | 13739000 | 303999 | 46 |
| Deletion | Chr3 | 14986928 | 14986929 | 1 | |
| Deletion | Chr2 | 15401597 | 15401607 | 10 | |
| Deletion | Chr11 | 15876188 | 15876203 | 15 | |
| Deletion | Chr10 | 20553548 | 20553549 | 1 | |
| Deletion | Chr3 | 24424899 | 24424900 | 1 | |
| Deletion | Chr2 | 25690118 | 25690132 | 14 | LOC_Os02g42720 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16889877 | 16889878 | 2 | |
| Insertion | Chr2 | 30953139 | 30953140 | 2 | |
| Insertion | Chr4 | 2102917 | 2102918 | 2 | |
| Insertion | Chr7 | 6945344 | 6945344 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 3360739 | Chr6 | 7698605 | 2 |
| Translocation | Chr12 | 6808362 | Chr11 | 13095610 |
