Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2533-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2533-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 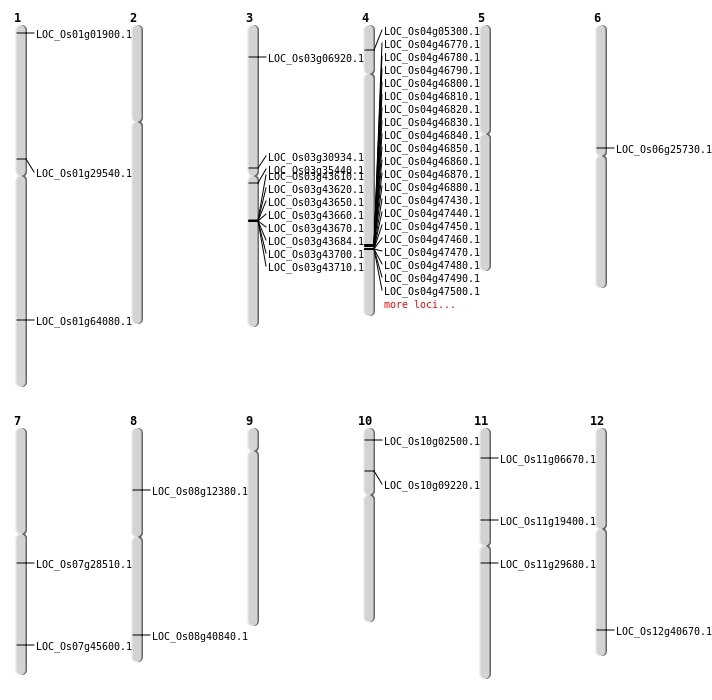
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10901815 | T-C | HET | ||
| SBS | Chr1 | 12152801 | C-A | HET | ||
| SBS | Chr1 | 19537433 | G-T | HET | ||
| SBS | Chr1 | 22415709 | C-T | HET | ||
| SBS | Chr1 | 41310523 | G-C | HET | ||
| SBS | Chr1 | 8039391 | T-C | HET | ||
| SBS | Chr10 | 13673727 | T-G | HET | ||
| SBS | Chr10 | 18131442 | C-T | HET | ||
| SBS | Chr10 | 19420732 | A-G | Homo | ||
| SBS | Chr10 | 19420734 | A-T | Homo | ||
| SBS | Chr10 | 240383 | A-G | HET | ||
| SBS | Chr10 | 4980890 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g09220 |
| SBS | Chr10 | 928181 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g02500 |
| SBS | Chr11 | 11157804 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g19400 |
| SBS | Chr11 | 16096940 | C-T | Homo | ||
| SBS | Chr11 | 17205727 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29680 |
| SBS | Chr11 | 28241693 | C-A | HET | ||
| SBS | Chr11 | 5539483 | A-T | HET | ||
| SBS | Chr12 | 13894541 | T-C | Homo | ||
| SBS | Chr2 | 23034896 | C-T | HET | ||
| SBS | Chr2 | 3610807 | G-A | HET | ||
| SBS | Chr2 | 3905820 | C-A | HET | ||
| SBS | Chr2 | 5765606 | C-T | HET | ||
| SBS | Chr2 | 8482121 | C-T | HET | ||
| SBS | Chr3 | 14027186 | C-A | Homo | ||
| SBS | Chr3 | 14027188 | T-C | Homo | ||
| SBS | Chr3 | 18157017 | T-C | Homo | ||
| SBS | Chr3 | 31724089 | T-C | HET | ||
| SBS | Chr3 | 32296529 | C-A | HET | ||
| SBS | Chr3 | 34405202 | C-T | HET | ||
| SBS | Chr3 | 4138620 | A-T | HET | ||
| SBS | Chr3 | 4973249 | C-T | HET | ||
| SBS | Chr3 | 8074730 | T-C | Homo | ||
| SBS | Chr4 | 3528276 | T-C | HET | ||
| SBS | Chr6 | 15027769 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25730 |
| SBS | Chr6 | 28099048 | T-A | HET | ||
| SBS | Chr7 | 13653932 | T-C | HET | ||
| SBS | Chr7 | 16686661 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28510 |
| SBS | Chr7 | 17845212 | T-C | HET | ||
| SBS | Chr7 | 27704451 | A-G | Homo | ||
| SBS | Chr8 | 12047315 | T-A | HET | ||
| SBS | Chr8 | 12065273 | G-A | HET | ||
| SBS | Chr8 | 25845642 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g40840 |
| SBS | Chr8 | 7295329 | T-A | HET | STOP_GAINED | LOC_Os08g12380 |
| SBS | Chr9 | 17997457 | G-A | HET | ||
| SBS | Chr9 | 20533222 | C-T | Homo | ||
| SBS | Chr9 | 594806 | G-A | Homo | ||
| SBS | Chr9 | 595997 | C-A | Homo |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 363924 | 363931 | 7 | |
| Deletion | Chr1 | 488959 | 488980 | 21 | LOC_Os01g01900 |
| Deletion | Chr4 | 7776750 | 7776752 | 2 | |
| Deletion | Chr7 | 10433963 | 10433965 | 2 | |
| Deletion | Chr1 | 11667817 | 11667910 | 93 | |
| Deletion | Chr3 | 14114765 | 14114774 | 9 | |
| Deletion | Chr3 | 19648163 | 19648164 | 1 | LOC_Os03g35440 |
| Deletion | Chr12 | 23436789 | 23436809 | 20 | |
| Deletion | Chr3 | 24370001 | 24447000 | 76999 | 8 |
| Deletion | Chr12 | 25179177 | 25179178 | 1 | LOC_Os12g40670 |
| Deletion | Chr4 | 27594280 | 27594281 | 1 | |
| Deletion | Chr4 | 27729001 | 27791000 | 61999 | 12 |
| Deletion | Chr4 | 28144001 | 28242000 | 97999 | 17 |
| Deletion | Chr4 | 28249001 | 28297000 | 47999 | 11 |
| Deletion | Chr4 | 30554765 | 30554766 | 1 | |
| Deletion | Chr1 | 37224333 | 37224334 | 1 | LOC_Os01g64080 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 9229235 | 9229235 | 1 | |
| Insertion | Chr2 | 23150804 | 23150806 | 3 | |
| Insertion | Chr9 | 3141605 | 3141605 | 1 |
Inversions: 6
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3249472 | Chr1 | 2526286 | LOC_Os11g06670 |
| Translocation | Chr8 | 8339347 | Chr7 | 3172532 | |
| Translocation | Chr2 | 16887664 | Chr1 | 16560201 | 2 |
| Translocation | Chr11 | 17928556 | Chr9 | 5981935 | |
| Translocation | Chr7 | 18351899 | Chr3 | 3505771 | 2 |
| Translocation | Chr5 | 18985805 | Chr2 | 3226637 | |
| Translocation | Chr4 | 19146632 | Chr3 | 17621955 | 2 |
| Translocation | Chr7 | 27214486 | Chr4 | 2638843 | 2 |
