Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2535-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2535-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 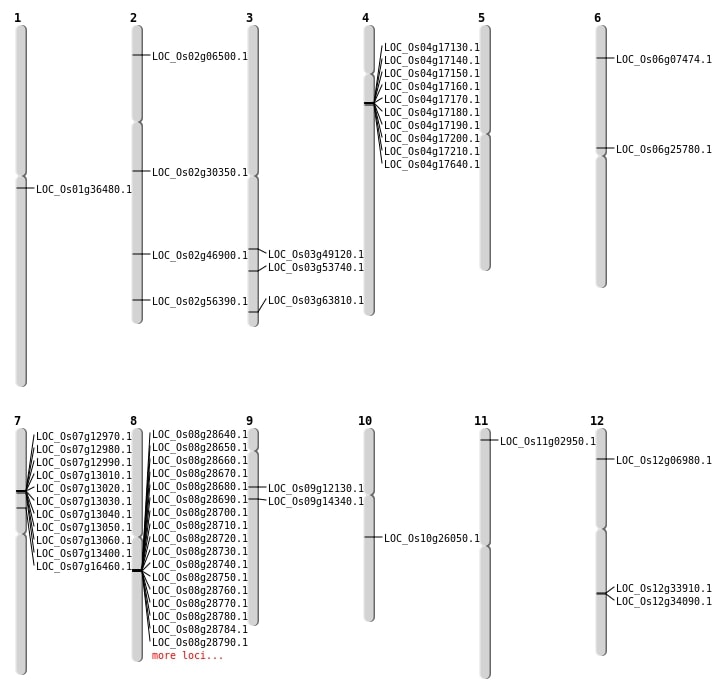
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13730501 | T-C | HET | ||
| SBS | Chr1 | 20249950 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g36480 |
| SBS | Chr1 | 23538371 | C-G | HET | ||
| SBS | Chr1 | 34268896 | C-T | HET | ||
| SBS | Chr1 | 37112877 | A-G | Homo | ||
| SBS | Chr1 | 43063476 | A-C | Homo | ||
| SBS | Chr1 | 43063477 | C-A | Homo | ||
| SBS | Chr10 | 13495400 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26050 |
| SBS | Chr10 | 19209662 | A-T | HET | ||
| SBS | Chr10 | 21529600 | T-C | HET | ||
| SBS | Chr10 | 6762971 | C-G | HET | ||
| SBS | Chr10 | 8000369 | T-C | HET | ||
| SBS | Chr11 | 17732187 | C-T | HET | ||
| SBS | Chr12 | 20437607 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g33910 |
| SBS | Chr12 | 20596145 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g34090 |
| SBS | Chr12 | 20596146 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g34090 |
| SBS | Chr12 | 20596147 | C-T | Homo | ||
| SBS | Chr12 | 3411286 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06980 |
| SBS | Chr12 | 9424739 | C-T | Homo | ||
| SBS | Chr2 | 11099418 | G-A | HET | ||
| SBS | Chr2 | 11099881 | A-T | HET | ||
| SBS | Chr2 | 2182351 | A-G | HET | ||
| SBS | Chr2 | 28611472 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g46900 |
| SBS | Chr2 | 30660980 | C-A | HET | ||
| SBS | Chr2 | 3239853 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g06500 |
| SBS | Chr3 | 27981856 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g49120 |
| SBS | Chr3 | 35329786 | G-A | HET | ||
| SBS | Chr3 | 8979471 | G-A | HET | ||
| SBS | Chr4 | 1445771 | G-C | HET | ||
| SBS | Chr4 | 14533348 | T-C | HET | ||
| SBS | Chr4 | 25814028 | A-G | HET | ||
| SBS | Chr4 | 6269660 | T-C | HET | ||
| SBS | Chr4 | 7780983 | A-G | HET | ||
| SBS | Chr4 | 9660456 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17640 |
| SBS | Chr5 | 462628 | A-G | HET | ||
| SBS | Chr7 | 15068589 | C-T | HET | ||
| SBS | Chr7 | 23056696 | A-G | HET | ||
| SBS | Chr7 | 8404042 | C-T | HET | ||
| SBS | Chr7 | 9195249 | C-A | HET | ||
| SBS | Chr7 | 9645562 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16460 |
| SBS | Chr7 | 9645563 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16460 |
| SBS | Chr8 | 18116882 | G-C | Homo | ||
| SBS | Chr9 | 15124765 | C-T | HET | ||
| SBS | Chr9 | 21196033 | A-G | HET | ||
| SBS | Chr9 | 21904232 | C-T | HET | ||
| SBS | Chr9 | 22552301 | G-T | HET | ||
| SBS | Chr9 | 6719864 | T-C | HET | ||
| SBS | Chr9 | 6859100 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g12130 |
| SBS | Chr9 | 7299822 | T-C | HET |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 64116 | 64470 | 354 | |
| Deletion | Chr9 | 120424 | 120425 | 1 | |
| Deletion | Chr4 | 7071480 | 7071481 | 1 | |
| Deletion | Chr4 | 7219471 | 7219478 | 7 | |
| Deletion | Chr7 | 7439001 | 7491000 | 51999 | 9 |
| Deletion | Chr4 | 9382001 | 9436000 | 53999 | 9 |
| Deletion | Chr12 | 10641914 | 10641922 | 8 | |
| Deletion | Chr11 | 11380109 | 11380110 | 1 | |
| Deletion | Chr6 | 15060394 | 15060402 | 8 | LOC_Os06g25780 |
| Deletion | Chr8 | 17497001 | 17739000 | 241999 | 36 |
| Deletion | Chr8 | 17745001 | 17873000 | 127999 | 17 |
| Deletion | Chr8 | 18563001 | 18825000 | 261999 | 34 |
| Deletion | Chr5 | 20834674 | 20834677 | 3 | |
| Deletion | Chr11 | 28824196 | 28824210 | 14 | |
| Deletion | Chr3 | 34276544 | 34276551 | 7 | |
| Deletion | Chr2 | 34523282 | 34523314 | 32 | LOC_Os02g56390 |
| Deletion | Chr3 | 36030001 | 36043000 | 12999 | LOC_Os03g63810 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 3391997 | 3392003 | 7 |
Inversions: 5
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 3595670 | Chr1 | 12316488 | LOC_Os06g07474 |
| Translocation | Chr6 | 3700326 | Chr1 | 12316543 | |
| Translocation | Chr6 | 7292234 | Chr2 | 18076972 | 2 |
| Translocation | Chr7 | 7439158 | Chr3 | 30809728 | 2 |
| Translocation | Chr7 | 7441341 | Chr3 | 30809733 | 2 |
| Translocation | Chr7 | 7689117 | Chr1 | 12316565 | LOC_Os07g13400 |
| Translocation | Chr9 | 8475172 | Chr5 | 8030676 | LOC_Os09g14340 |
| Translocation | Chr4 | 9382368 | Chr2 | 28251277 | LOC_Os04g17130 |
