Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2540-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2540-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 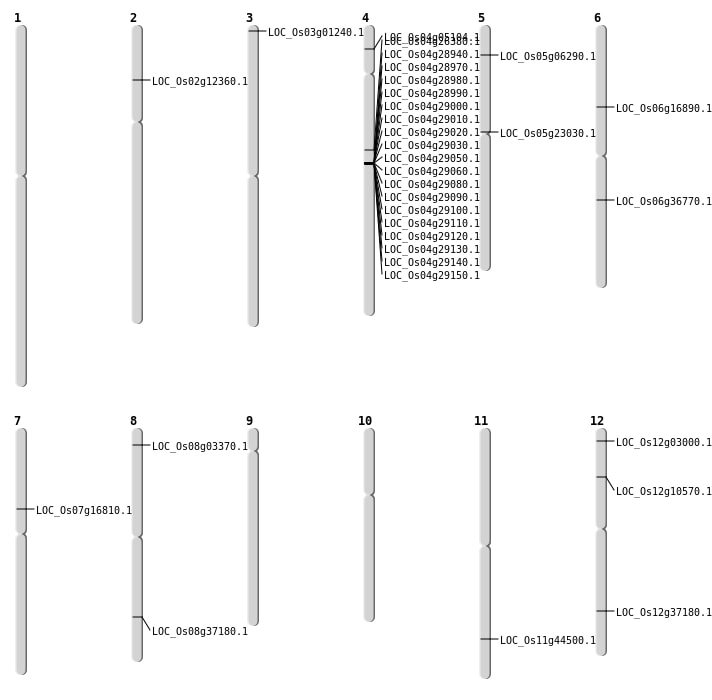
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12013243 | C-T | HET | ||
| SBS | Chr1 | 2258601 | A-G | HET | ||
| SBS | Chr1 | 2922041 | A-G | Homo | ||
| SBS | Chr1 | 6442797 | G-A | Homo | ||
| SBS | Chr10 | 16488963 | T-A | HET | ||
| SBS | Chr10 | 1678099 | A-G | Homo | ||
| SBS | Chr10 | 22510643 | C-T | HET | ||
| SBS | Chr10 | 6125388 | C-T | Homo | ||
| SBS | Chr11 | 23795746 | G-A | HET | ||
| SBS | Chr11 | 26917545 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44500 |
| SBS | Chr11 | 3019607 | C-T | Homo | ||
| SBS | Chr11 | 5306265 | T-G | HET | ||
| SBS | Chr11 | 6252719 | C-T | Homo | ||
| SBS | Chr12 | 1118422 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g03000 |
| SBS | Chr12 | 12612167 | T-G | HET | ||
| SBS | Chr12 | 24609490 | C-T | HET | ||
| SBS | Chr12 | 24921888 | G-T | HET | ||
| SBS | Chr2 | 3392122 | A-G | HET | ||
| SBS | Chr2 | 5455761 | G-A | HET | ||
| SBS | Chr2 | 6435676 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g12360 |
| SBS | Chr2 | 7132079 | G-T | HET | ||
| SBS | Chr3 | 19102785 | G-T | Homo | ||
| SBS | Chr3 | 31095530 | T-A | HET | ||
| SBS | Chr3 | 6100962 | T-C | HET | ||
| SBS | Chr3 | 8848736 | C-T | HET | ||
| SBS | Chr4 | 12492036 | T-C | HET | ||
| SBS | Chr4 | 15388901 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g26380 |
| SBS | Chr4 | 15922993 | G-T | HET | ||
| SBS | Chr4 | 17336572 | C-T | Homo | ||
| SBS | Chr4 | 2523975 | C-A | HET | STOP_GAINED | LOC_Os04g05104 |
| SBS | Chr5 | 14776518 | G-A | HET | ||
| SBS | Chr5 | 3216183 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g06290 |
| SBS | Chr5 | 9376747 | T-A | HET | ||
| SBS | Chr6 | 13857441 | T-C | HET | ||
| SBS | Chr6 | 13857442 | T-A | HET | ||
| SBS | Chr6 | 1684486 | C-A | Homo | ||
| SBS | Chr6 | 8148031 | T-C | HET | ||
| SBS | Chr6 | 9792388 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g16890 |
| SBS | Chr7 | 10464944 | G-A | HET | ||
| SBS | Chr7 | 13633802 | C-A | HET | ||
| SBS | Chr7 | 8563563 | A-G | HET | ||
| SBS | Chr7 | 9866506 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16810 |
| SBS | Chr8 | 23494056 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g37180 |
| SBS | Chr9 | 10149997 | A-C | Homo | ||
| SBS | Chr9 | 20255881 | C-T | HET | ||
| SBS | Chr9 | 8755341 | C-A | HET | ||
| SBS | Chr9 | 8755342 | C-T | HET |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 5248533 | 5248541 | 8 | |
| Deletion | Chr12 | 5612246 | 5612251 | 5 | LOC_Os12g10570 |
| Deletion | Chr5 | 7567508 | 7567523 | 15 | |
| Deletion | Chr5 | 7828412 | 7828439 | 27 | |
| Deletion | Chr6 | 8832782 | 8832788 | 6 | |
| Deletion | Chr8 | 9223071 | 9223074 | 3 | |
| Deletion | Chr5 | 13132876 | 13132880 | 4 | LOC_Os05g23030 |
| Deletion | Chr3 | 13452684 | 13452706 | 22 | |
| Deletion | Chr1 | 16545929 | 16545930 | 1 | |
| Deletion | Chr4 | 17157001 | 17205000 | 47999 | 8 |
| Deletion | Chr4 | 17210001 | 17289000 | 78999 | 10 |
| Deletion | Chr7 | 18969818 | 18969820 | 2 | |
| Deletion | Chr6 | 19413931 | 19413941 | 10 | |
| Deletion | Chr3 | 21602979 | 21603006 | 27 | |
| Deletion | Chr1 | 25023643 | 25023658 | 15 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 12834032 | 12834033 | 2 | |
| Insertion | Chr4 | 15065348 | 15065348 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 30640429 | 30640587 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 155198 | Chr1 | 5547586 | LOC_Os03g01240 |
| Translocation | Chr3 | 155782 | Chr1 | 5547624 | LOC_Os03g01240 |
| Translocation | Chr8 | 1578727 | Chr4 | 6811822 | LOC_Os08g03370 |
| Translocation | Chr8 | 1578728 | Chr4 | 6812774 | LOC_Os08g03370 |
| Translocation | Chr9 | 4757267 | Chr4 | 1178866 | |
| Translocation | Chr6 | 21657463 | Chr4 | 17923880 | LOC_Os06g36770 |
| Translocation | Chr12 | 22800140 | Chr9 | 4757324 | LOC_Os12g37180 |
