Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3067-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3067-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 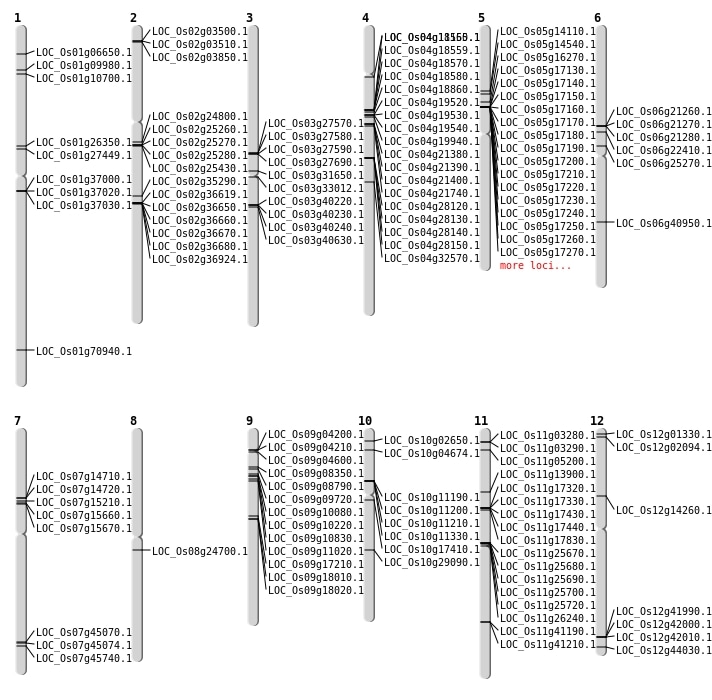
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr11 | 14801621 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 15897955 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 11485531 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 19722859 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 22018408 | C-A | HET | INTRON | |
| SBS | Chr3 | 25084805 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 27994527 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 34494609 | A-G | HET | INTRON | |
| SBS | Chr3 | 7911689 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21721511 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30952304 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 10480302 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 133468 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16690296 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g28500 |
| SBS | Chr5 | 24451797 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 24608300 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 5082164 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 7229018 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 20326483 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 20691261 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr7 | 23619309 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 14927390 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g24700 |
| SBS | Chr8 | 5194750 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 5529014 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 10294710 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 2582478 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 3297420 | G-A | HET | INTERGENIC |
Deletions: 87
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 179001 | 190000 | 10999 | 2 |
| Deletion | Chr10 | 1022001 | 1039000 | 16999 | 2 |
| Deletion | Chr11 | 1203001 | 1227000 | 23999 | 3 |
| Deletion | Chr2 | 1418001 | 1429000 | 10999 | 3 |
| Deletion | Chr3 | 1959977 | 1959980 | 3 | |
| Deletion | Chr9 | 2203001 | 2217000 | 13999 | 3 |
| Deletion | Chr4 | 2863397 | 2863412 | 15 | |
| Deletion | Chr9 | 4311001 | 4327000 | 15999 | 2 |
| Deletion | Chr1 | 5189001 | 5201000 | 11999 | 2 |
| Deletion | Chr9 | 5260299 | 5260300 | 1 | LOC_Os09g09720 |
| Deletion | Chr9 | 5480001 | 5493000 | 12999 | 2 |
| Deletion | Chr10 | 5513261 | 5513262 | 1 | |
| Deletion | Chr4 | 5615001 | 5629000 | 13999 | LOC_Os04g11165 |
| Deletion | Chr9 | 5768749 | 5768767 | 18 | |
| Deletion | Chr9 | 5903001 | 5912000 | 8999 | 2 |
| Deletion | Chr7 | 6160026 | 6160027 | 1 | |
| Deletion | Chr10 | 6198001 | 6221000 | 22999 | 4 |
| Deletion | Chr11 | 7599001 | 7612000 | 12999 | LOC_Os11g13900 |
| Deletion | Chr9 | 7607268 | 7607269 | 1 | |
| Deletion | Chr12 | 7626001 | 7637000 | 10999 | LOC_Os12g14260 |
| Deletion | Chr5 | 7873001 | 7888000 | 14999 | 2 |
| Deletion | Chr7 | 8387001 | 8402000 | 14999 | 2 |
| Deletion | Chr4 | 8953683 | 8953738 | 55 | |
| Deletion | Chr7 | 9086001 | 9100000 | 13999 | 2 |
| Deletion | Chr7 | 9167236 | 9167237 | 1 | |
| Deletion | Chr5 | 9197001 | 9214000 | 16999 | 2 |
| Deletion | Chr1 | 9548001 | 9560000 | 11999 | |
| Deletion | Chr11 | 9656001 | 9669000 | 12999 | 2 |
| Deletion | Chr11 | 9712001 | 9726000 | 13999 | 3 |
| Deletion | Chr5 | 9786001 | 10016000 | 229999 | 33 |
| Deletion | Chr5 | 10021001 | 10044000 | 22999 | 2 |
| Deletion | Chr5 | 10058001 | 10189000 | 130999 | 17 |
| Deletion | Chr5 | 10201001 | 10373000 | 171999 | 27 |
| Deletion | Chr4 | 10244001 | 10266000 | 21999 | 5 |
| Deletion | Chr5 | 10300460 | 10300461 | 1 | |
| Deletion | Chr5 | 10371168 | 10371169 | 1 | |
| Deletion | Chr5 | 10376001 | 10526000 | 149999 | 24 |
| Deletion | Chr5 | 10630001 | 10679000 | 48999 | 6 |
| Deletion | Chr5 | 10649631 | 10649632 | 1 | |
| Deletion | Chr4 | 10862001 | 10878000 | 15999 | 4 |
| Deletion | Chr9 | 11026001 | 11053000 | 26999 | 2 |
| Deletion | Chr6 | 11245776 | 11245777 | 1 | |
| Deletion | Chr4 | 12071001 | 12092000 | 20999 | 4 |
| Deletion | Chr6 | 12287001 | 12303000 | 15999 | 4 |
| Deletion | Chr5 | 12628001 | 12660000 | 31999 | 3 |
| Deletion | Chr5 | 12663001 | 12679000 | 15999 | 2 |
| Deletion | Chr10 | 13223458 | 13223469 | 11 | |
| Deletion | Chr12 | 13377146 | 13377231 | 85 | |
| Deletion | Chr9 | 13930097 | 13930102 | 5 | |
| Deletion | Chr1 | 14028969 | 14028970 | 1 | |
| Deletion | Chr6 | 14229001 | 14241000 | 11999 | |
| Deletion | Chr2 | 14391395 | 14391397 | 2 | LOC_Os02g24800 |
| Deletion | Chr9 | 14504962 | 14504967 | 5 | |
| Deletion | Chr11 | 14641001 | 14664000 | 22999 | 6 |
| Deletion | Chr2 | 14700001 | 14731000 | 30999 | 4 |
| Deletion | Chr6 | 14781485 | 14781486 | 1 | LOC_Os06g25270 |
| Deletion | Chr1 | 14939001 | 14949000 | 9999 | 2 |
| Deletion | Chr3 | 15805001 | 15824000 | 18999 | 4 |
| Deletion | Chr1 | 15894423 | 15894424 | 1 | |
| Deletion | Chr4 | 16597001 | 16626000 | 28999 | 4 |
| Deletion | Chr10 | 17340129 | 17340139 | 10 | |
| Deletion | Chr4 | 17451027 | 17451091 | 64 | |
| Deletion | Chr3 | 18069001 | 18079000 | 9999 | 2 |
| Deletion | Chr1 | 18899009 | 18899010 | 1 | |
| Deletion | Chr4 | 19608791 | 19608795 | 4 | LOC_Os04g32570 |
| Deletion | Chr11 | 20488457 | 20488460 | 3 | |
| Deletion | Chr1 | 20636001 | 20659000 | 22999 | 3 |
| Deletion | Chr2 | 21204001 | 21214000 | 9999 | 2 |
| Deletion | Chr10 | 21537399 | 21537407 | 8 | |
| Deletion | Chr2 | 22118001 | 22141000 | 22999 | 5 |
| Deletion | Chr3 | 22353001 | 22366000 | 12999 | 4 |
| Deletion | Chr8 | 22727921 | 22727923 | 2 | |
| Deletion | Chr6 | 24415001 | 24429000 | 13999 | LOC_Os06g40950 |
| Deletion | Chr11 | 24677001 | 24698000 | 20999 | 2 |
| Deletion | Chr8 | 24923957 | 24923962 | 5 | |
| Deletion | Chr11 | 25740024 | 25740027 | 3 | |
| Deletion | Chr12 | 26032001 | 26047000 | 14999 | 4 |
| Deletion | Chr3 | 26243809 | 26243811 | 2 | |
| Deletion | Chr7 | 26891001 | 26904000 | 12999 | 3 |
| Deletion | Chr8 | 28282533 | 28282537 | 4 | |
| Deletion | Chr6 | 29461484 | 29461487 | 3 | |
| Deletion | Chr3 | 30647832 | 30647838 | 6 | |
| Deletion | Chr3 | 30647846 | 30647852 | 6 | |
| Deletion | Chr1 | 32342484 | 32342485 | 1 | |
| Deletion | Chr4 | 33293328 | 33293370 | 42 | |
| Deletion | Chr1 | 38652064 | 38652072 | 8 | |
| Deletion | Chr1 | 40346001 | 40366000 | 19999 | LOC_Os01g70940 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21005996 | 21005996 | 1 | |
| Insertion | Chr10 | 4486016 | 4486016 | 1 | |
| Insertion | Chr11 | 4650373 | 4650382 | 10 | |
| Insertion | Chr12 | 12616516 | 12616516 | 1 | |
| Insertion | Chr12 | 22116317 | 22116320 | 4 | |
| Insertion | Chr4 | 14679624 | 14679625 | 2 | |
| Insertion | Chr4 | 4021831 | 4021831 | 1 | |
| Insertion | Chr5 | 10292886 | 10292886 | 1 | |
| Insertion | Chr5 | 10317626 | 10317626 | 1 | |
| Insertion | Chr5 | 10317829 | 10317829 | 1 | |
| Insertion | Chr5 | 10431881 | 10431881 | 1 | |
| Insertion | Chr6 | 28707105 | 28707105 | 1 | |
| Insertion | Chr7 | 17734106 | 17734106 | 1 | |
| Insertion | Chr7 | 8766718 | 8766718 | 1 | LOC_Os07g15210 |
| Insertion | Chr7 | 9892461 | 9892461 | 1 | |
| Insertion | Chr8 | 12442705 | 12442705 | 1 | |
| Insertion | Chr8 | 6625934 | 6625934 | 1 | |
| Insertion | Chr8 | 8547102 | 8547102 | 1 | |
| Insertion | Chr9 | 12056059 | 12056059 | 1 | |
| Insertion | Chr9 | 5920546 | 5920547 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 34597026 | 35113061 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 516904 | Chr7 | 11770274 | |
| Translocation | Chr12 | 2005847 | Chr2 | 19681000 | |
| Translocation | Chr12 | 6229548 | Chr2 | 27057759 | |
| Translocation | Chr11 | 8196161 | Chr7 | 11141499 | |
| Translocation | Chr10 | 8778262 | Chr9 | 10566383 | 2 |
| Translocation | Chr8 | 14757240 | Chr2 | 31803319 | |
| Translocation | Chr8 | 14757409 | Chr2 | 31803320 | |
| Translocation | Chr10 | 15151363 | Chr5 | 26317302 | LOC_Os10g29090 |
| Translocation | Chr10 | 15151378 | Chr1 | 3134836 | 2 |
| Translocation | Chr12 | 17347650 | Chr4 | 2085444 | |
| Translocation | Chr8 | 19434238 | Chr6 | 494463 | |
| Translocation | Chr3 | 22753954 | Chr1 | 37534642 | |
| Translocation | Chr3 | 22753970 | Chr1 | 38541764 | |
| Translocation | Chr11 | 24593816 | Chr6 | 26017510 |
