Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN369-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN369-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 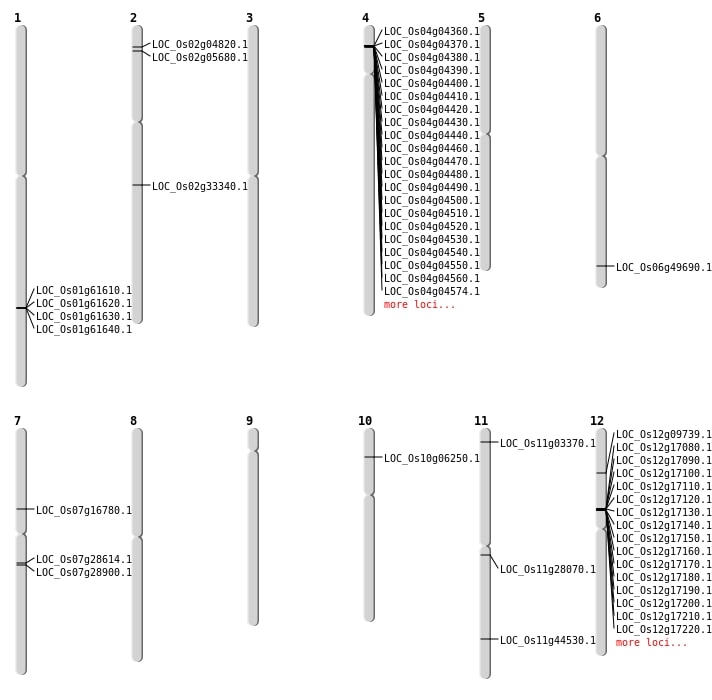
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 7083788 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 3206423 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g06250 |
| SBS | Chr11 | 10526323 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12796622 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 9415292 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 10034894 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 16566157 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 18431894 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 18804700 | A-T | HOMO | INTRON | |
| SBS | Chr12 | 24242495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 26402118 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 13485243 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 19811708 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g33340 |
| SBS | Chr2 | 27936918 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 27936924 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7189071 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 21399439 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27362661 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 30194412 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 9863238 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 9898730 | A-G | HET | INTRON | |
| SBS | Chr5 | 10520036 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 14200338 | T-G | HET | INTRON | |
| SBS | Chr5 | 3830368 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 15066808 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 1065828 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 16749123 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28614 |
| SBS | Chr7 | 16946291 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28900 |
| SBS | Chr8 | 23987722 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12081052 | T-C | HET | INTRON | |
| SBS | Chr9 | 9120978 | G-A | HET | INTRON |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1325723 | 1325733 | 10 | |
| Deletion | Chr4 | 2050001 | 2265000 | 215000 | 33 |
| Deletion | Chr2 | 2309783 | 2309784 | 1 | |
| Deletion | Chr12 | 6926405 | 6926417 | 12 | |
| Deletion | Chr12 | 9780001 | 9999000 | 219000 | 37 |
| Deletion | Chr7 | 9849918 | 9849923 | 5 | LOC_Os07g16780 |
| Deletion | Chr6 | 10776806 | 10776815 | 9 | |
| Deletion | Chr2 | 11218389 | 11218390 | 1 | |
| Deletion | Chr10 | 13946149 | 13946167 | 18 | |
| Deletion | Chr9 | 14705826 | 14705828 | 2 | |
| Deletion | Chr5 | 15257521 | 15257538 | 17 | |
| Deletion | Chr11 | 16137483 | 16137489 | 6 | LOC_Os11g28070 |
| Deletion | Chr12 | 18804177 | 18804194 | 17 | |
| Deletion | Chr10 | 18950549 | 18950550 | 1 | |
| Deletion | Chr8 | 23931351 | 23931358 | 7 | |
| Deletion | Chr3 | 24411460 | 24411470 | 10 | |
| Deletion | Chr6 | 26575902 | 26575910 | 8 | |
| Deletion | Chr11 | 26929026 | 26929027 | 1 | LOC_Os11g44530 |
| Deletion | Chr5 | 27405988 | 27405998 | 10 | |
| Deletion | Chr5 | 27798790 | 27798791 | 1 | |
| Deletion | Chr5 | 28370325 | 28370337 | 12 | |
| Deletion | Chr6 | 30066308 | 30066313 | 5 | LOC_Os06g49690 |
| Deletion | Chr1 | 35633897 | 35663588 | 29691 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21109752 | 21109752 | 1 | |
| Insertion | Chr1 | 27782763 | 27782764 | 2 | |
| Insertion | Chr3 | 3312217 | 3312217 | 1 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 653372 | 2050060 | |
| Inversion | Chr2 | 2223974 | 2777995 | 2 |
| Inversion | Chr2 | 2223988 | 2778000 | 2 |
| Inversion | Chr12 | 5143181 | 6229526 | LOC_Os12g09739 |
| Inversion | Chr12 | 9779744 | 10277549 | LOC_Os12g17080 |
| Inversion | Chr12 | 9999384 | 10277549 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5143224 | Chr11 | 1270190 | 2 |
